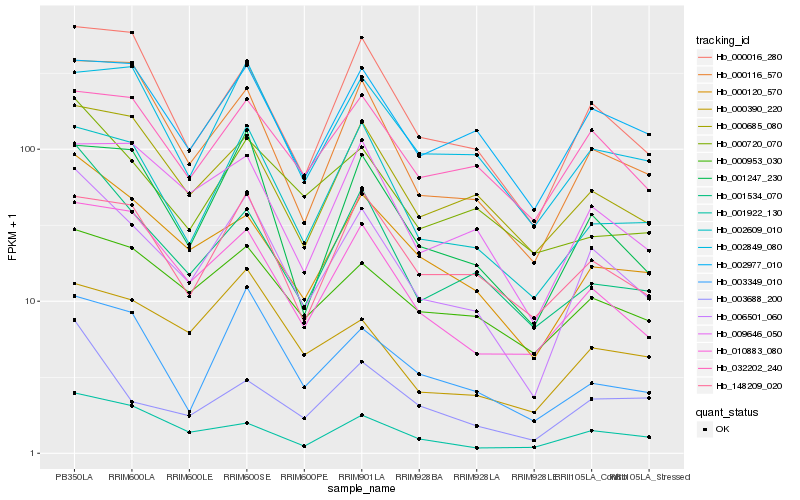
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006501_060 |
0.0 |
- |
- |
PREDICTED: lachrymatory-factor synthase-like [Jatropha curcas] |
| 2 |
Hb_000120_570 |
0.1232578028 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 3 |
Hb_002609_010 |
0.1325557897 |
- |
- |
PREDICTED: uncharacterized protein LOC105630866 [Jatropha curcas] |
| 4 |
Hb_001247_230 |
0.1338876053 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_010883_080 |
0.1341829881 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001534_070 |
0.134299531 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Eucalyptus grandis] |
| 7 |
Hb_000116_570 |
0.1371549843 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 8 |
Hb_000016_280 |
0.1389721205 |
- |
- |
Arginine/serine-rich splicing factor 35 [Theobroma cacao] |
| 9 |
Hb_000685_080 |
0.1433585138 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_002977_010 |
0.1459084151 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 11 |
Hb_000953_030 |
0.1462370353 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_003349_010 |
0.1475127599 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CICLE_v10023911mg [Citrus clementina] |
| 13 |
Hb_000720_070 |
0.1476442644 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_20639 [Jatropha curcas] |
| 14 |
Hb_148209_020 |
0.1492167516 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 15 |
Hb_001922_130 |
0.150504694 |
- |
- |
hypothetical protein PHAVU_002G294300g [Phaseolus vulgaris] |
| 16 |
Hb_032202_240 |
0.1510267314 |
- |
- |
PREDICTED: F-box protein At4g00755 [Jatropha curcas] |
| 17 |
Hb_009646_050 |
0.1523417092 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000390_220 |
0.1531921437 |
- |
- |
PREDICTED: uncharacterized protein LOC100244219 [Vitis vinifera] |
| 19 |
Hb_002849_080 |
0.1534564764 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 20 |
Hb_003688_200 |
0.1555643229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |