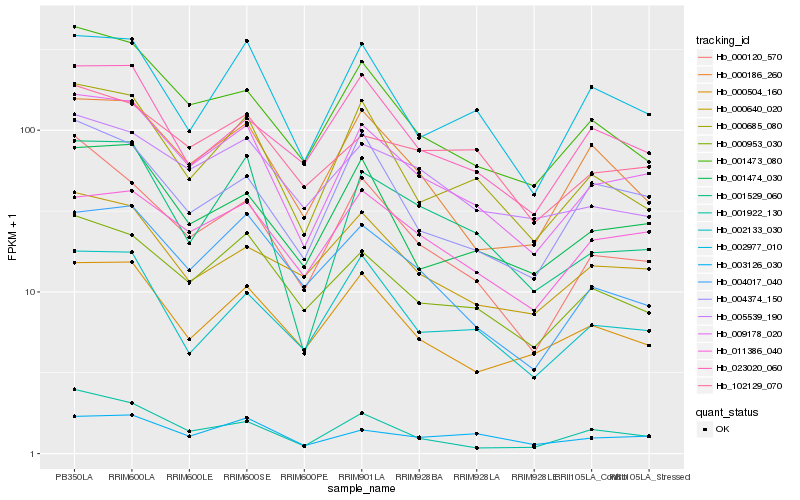
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009178_020 |
0.0 |
- |
- |
PREDICTED: zinc finger protein 622 [Jatropha curcas] |
| 2 |
Hb_000953_030 |
0.0903968988 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_001474_030 |
0.09466073 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 4 |
Hb_001473_080 |
0.0950788543 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 5 |
Hb_000685_080 |
0.095428837 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_000504_160 |
0.101194906 |
- |
- |
hypothetical protein POPTR_0001s35340g [Populus trichocarpa] |
| 7 |
Hb_000186_260 |
0.1020982232 |
transcription factor |
TF Family: PHD |
PREDICTED: protein polybromo-1-like [Jatropha curcas] |
| 8 |
Hb_003126_030 |
0.1027698685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001529_060 |
0.1034209271 |
- |
- |
PREDICTED: U4/U6.U5 small nuclear ribonucleoprotein 27 kDa protein [Jatropha curcas] |
| 10 |
Hb_000120_570 |
0.1064884514 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 11 |
Hb_002977_010 |
0.1066723666 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 12 |
Hb_011386_040 |
0.107535144 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004017_040 |
0.1082863522 |
- |
- |
kinetochore protein nuf2, putative [Ricinus communis] |
| 14 |
Hb_102129_070 |
0.1085152341 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 124 [Cucumis melo] |
| 15 |
Hb_004374_150 |
0.108823769 |
- |
- |
PREDICTED: uncharacterized protein LOC105650290 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000640_020 |
0.1100802795 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 17 |
Hb_001922_130 |
0.1101077916 |
- |
- |
hypothetical protein PHAVU_002G294300g [Phaseolus vulgaris] |
| 18 |
Hb_002133_030 |
0.1103007498 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 1 [Jatropha curcas] |
| 19 |
Hb_023020_060 |
0.1104962969 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 20 |
Hb_005539_190 |
0.1105690009 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |