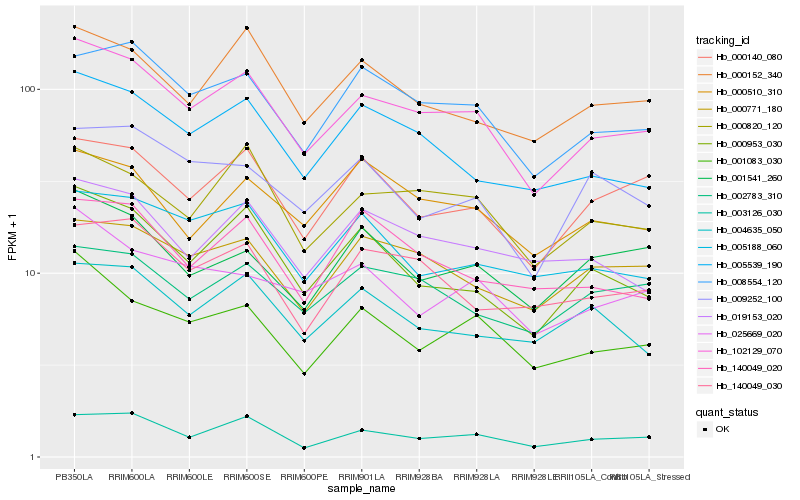
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_102129_070 |
0.0 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 124 [Cucumis melo] |
| 2 |
Hb_003126_030 |
0.0733633238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008554_120 |
0.0750216341 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 4 |
Hb_000820_120 |
0.0824488269 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 5 |
Hb_005539_190 |
0.0830956406 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 6 |
Hb_001083_030 |
0.0834655913 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase complex SCF subunit sconB [Jatropha curcas] |
| 7 |
Hb_000953_030 |
0.0847117661 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_000152_340 |
0.0897658208 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 9 |
Hb_000140_080 |
0.0928544423 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_019153_020 |
0.0947610485 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000771_180 |
0.0970250201 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 12 |
Hb_140049_030 |
0.097956919 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 13 |
Hb_005188_060 |
0.0989820483 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 14 |
Hb_025669_020 |
0.101007304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_140049_020 |
0.1012342039 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 16 |
Hb_000510_310 |
0.1024347559 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 17 |
Hb_002783_310 |
0.1034469251 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit trm6 [Jatropha curcas] |
| 18 |
Hb_001541_260 |
0.1038072938 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 19 |
Hb_004635_050 |
0.1040417063 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 20 |
Hb_009252_100 |
0.1040748281 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |