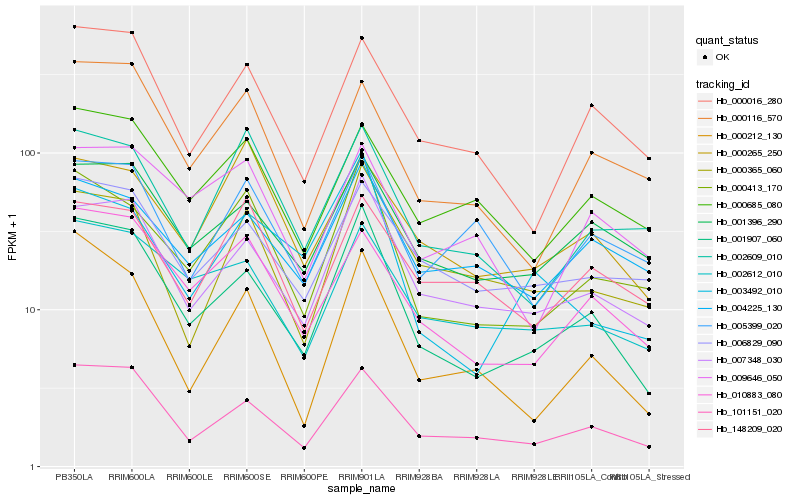
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000413_170 |
0.0 |
- |
- |
PREDICTED: probable histone H2A.3 [Jatropha curcas] |
| 2 |
Hb_002609_010 |
0.1009619303 |
- |
- |
PREDICTED: uncharacterized protein LOC105630866 [Jatropha curcas] |
| 3 |
Hb_001907_060 |
0.124996755 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 5 [Jatropha curcas] |
| 4 |
Hb_007348_030 |
0.1357628275 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004225_130 |
0.135968965 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105628060 isoform X1 [Jatropha curcas] |
| 6 |
Hb_148209_020 |
0.1422465774 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 7 |
Hb_010883_080 |
0.1452676713 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_101151_020 |
0.1473372504 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_002612_010 |
0.1480899241 |
- |
- |
hypothetical protein JCGZ_19269 [Jatropha curcas] |
| 10 |
Hb_000116_570 |
0.1481434048 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 11 |
Hb_000016_280 |
0.14834324 |
- |
- |
Arginine/serine-rich splicing factor 35 [Theobroma cacao] |
| 12 |
Hb_000365_060 |
0.1502338131 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 13 |
Hb_001396_290 |
0.1503450177 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 14 |
Hb_009646_050 |
0.1507795043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000685_080 |
0.150903723 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_000265_250 |
0.1519293066 |
- |
- |
auxilin, putative [Ricinus communis] |
| 17 |
Hb_006829_090 |
0.1530568044 |
- |
- |
PREDICTED: protein MOS2 [Jatropha curcas] |
| 18 |
Hb_000212_130 |
0.1551720022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003492_010 |
0.1558493381 |
- |
- |
DREPP plasma membrane polypeptide family protein [Populus trichocarpa] |
| 20 |
Hb_005399_020 |
0.1573863896 |
- |
- |
hypothetical protein CISIN_1g022820mg [Citrus sinensis] |