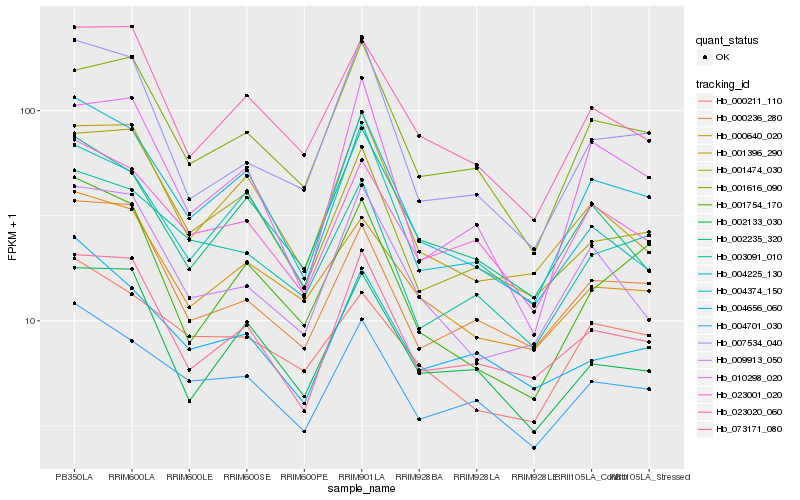
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004374_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650290 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004701_030 |
0.0805253717 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 3 |
Hb_073171_080 |
0.0826161451 |
- |
- |
PREDICTED: MMS19 nucleotide excision repair protein homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_023020_060 |
0.0832156629 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 5 |
Hb_001396_290 |
0.0847828381 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 6 |
Hb_023001_020 |
0.0854887642 |
- |
- |
PREDICTED: mitochondrial succinate-fumarate transporter 1 [Jatropha curcas] |
| 7 |
Hb_001474_030 |
0.0894775257 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 8 |
Hb_000640_020 |
0.0914699214 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 9 |
Hb_001754_170 |
0.0932803329 |
- |
- |
PREDICTED: 60S ribosome subunit biogenesis protein NIP7 homolog [Jatropha curcas] |
| 10 |
Hb_003091_010 |
0.0939044924 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 11 |
Hb_001616_090 |
0.0941708808 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF4 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004225_130 |
0.0943697037 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105628060 isoform X1 [Jatropha curcas] |
| 13 |
Hb_009913_050 |
0.09460288 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 14 |
Hb_000211_110 |
0.0950166343 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 15 |
Hb_000236_280 |
0.0958883278 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 22 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_002235_320 |
0.0970089803 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007534_040 |
0.0974450056 |
- |
- |
PREDICTED: uncharacterized protein LOC105635538 [Jatropha curcas] |
| 18 |
Hb_010298_020 |
0.097656857 |
- |
- |
PREDICTED: uncharacterized protein At2g02148 isoform X4 [Vitis vinifera] |
| 19 |
Hb_002133_030 |
0.0982103252 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 1 [Jatropha curcas] |
| 20 |
Hb_004656_060 |
0.1004443426 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |