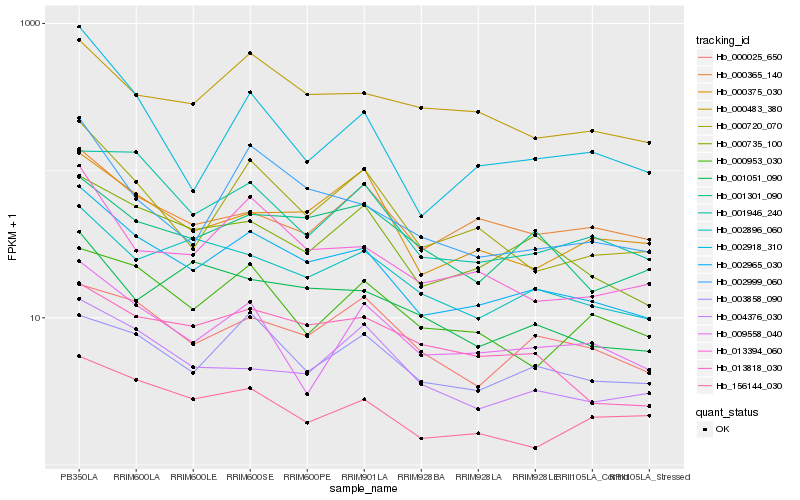
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002965_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 2 |
Hb_000735_100 |
0.1036549927 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 3 |
Hb_000720_070 |
0.1091780344 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_20639 [Jatropha curcas] |
| 4 |
Hb_013394_060 |
0.1092994126 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 5 |
Hb_002896_060 |
0.1206880133 |
- |
- |
PREDICTED: synaptotagmin-2 [Jatropha curcas] |
| 6 |
Hb_002999_060 |
0.1209821663 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 7 |
Hb_001946_240 |
0.1223464352 |
- |
- |
PREDICTED: chaperone protein dnaJ 49-like [Jatropha curcas] |
| 8 |
Hb_000483_380 |
0.128074699 |
- |
- |
PREDICTED: bax inhibitor 1-like [Jatropha curcas] |
| 9 |
Hb_001301_090 |
0.1286973915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_013818_030 |
0.1287178202 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003858_090 |
0.1294400393 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 12 |
Hb_000375_030 |
0.130806236 |
- |
- |
- |
| 13 |
Hb_004376_030 |
0.1322903048 |
transcription factor |
TF Family: mTERF |
hypothetical protein PHAVU_005G060900g [Phaseolus vulgaris] |
| 14 |
Hb_000025_650 |
0.1325025736 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 15 |
Hb_000365_140 |
0.1332812098 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase [Jatropha curcas] |
| 16 |
Hb_000953_030 |
0.1345027642 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_009558_040 |
0.1349161849 |
transcription factor |
TF Family: MYB |
hypothetical protein PHAVU_008G102000g [Phaseolus vulgaris] |
| 18 |
Hb_002918_310 |
0.1365220552 |
- |
- |
PREDICTED: dnaJ protein homolog 1-like [Jatropha curcas] |
| 19 |
Hb_156144_030 |
0.1365281977 |
- |
- |
hypothetical protein RCOM_0187400 [Ricinus communis] |
| 20 |
Hb_001051_090 |
0.1399341274 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |