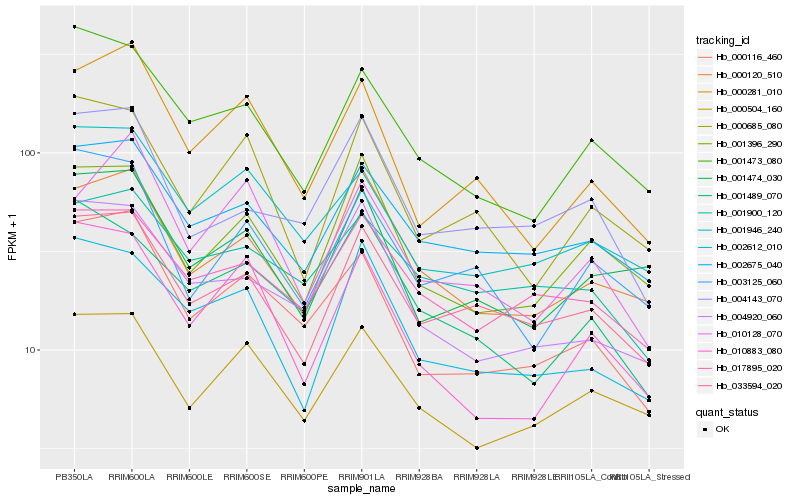
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_460 |
0.0 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 4 [Jatropha curcas] |
| 2 |
Hb_001946_240 |
0.0724014398 |
- |
- |
PREDICTED: chaperone protein dnaJ 49-like [Jatropha curcas] |
| 3 |
Hb_000281_010 |
0.0781392362 |
- |
- |
hypothetical protein JCGZ_16194 [Jatropha curcas] |
| 4 |
Hb_004920_060 |
0.1005479759 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 5 |
Hb_001473_080 |
0.1048301736 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 6 |
Hb_010883_080 |
0.108314535 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002675_040 |
0.1122138004 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 8 |
Hb_000504_160 |
0.1133542293 |
- |
- |
hypothetical protein POPTR_0001s35340g [Populus trichocarpa] |
| 9 |
Hb_003125_060 |
0.1187593804 |
- |
- |
PREDICTED: snurportin-1 [Jatropha curcas] |
| 10 |
Hb_033594_020 |
0.1192283737 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 11 |
Hb_004143_070 |
0.1215095028 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_001474_030 |
0.123365613 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 13 |
Hb_001900_120 |
0.1235243603 |
- |
- |
PREDICTED: uncharacterized protein LOC105632666 [Jatropha curcas] |
| 14 |
Hb_017895_020 |
0.1241751435 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 15 |
Hb_001489_070 |
0.12508454 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 16 |
Hb_010128_070 |
0.125643664 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_000685_080 |
0.1263147445 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_002612_010 |
0.1272595237 |
- |
- |
hypothetical protein JCGZ_19269 [Jatropha curcas] |
| 19 |
Hb_000120_510 |
0.1302659461 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001396_290 |
0.1306749487 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |