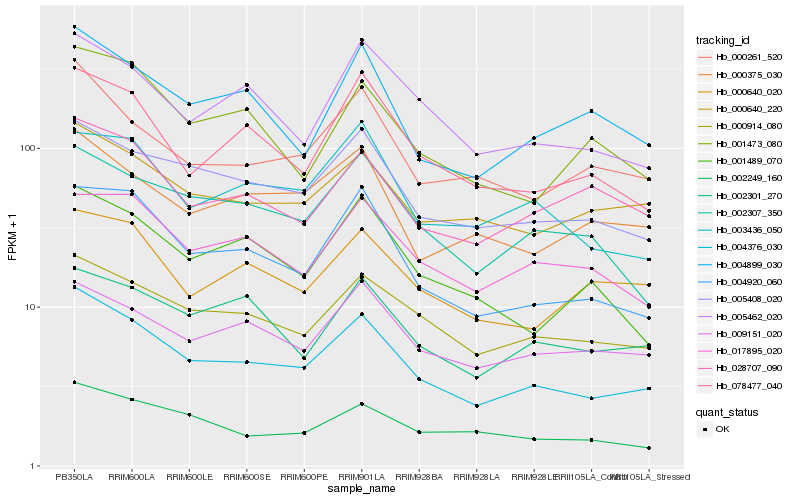
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004376_030 |
0.0 |
transcription factor |
TF Family: mTERF |
hypothetical protein PHAVU_005G060900g [Phaseolus vulgaris] |
| 2 |
Hb_005408_020 |
0.0844699359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000914_080 |
0.0887456533 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_004920_060 |
0.0891135664 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 5 |
Hb_000261_520 |
0.0955737751 |
- |
- |
PREDICTED: uncharacterized protein LOC105633192 [Jatropha curcas] |
| 6 |
Hb_000640_220 |
0.0966734302 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 7 |
Hb_078477_040 |
0.1031040466 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 8 |
Hb_005462_020 |
0.1040717908 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 9 |
Hb_002307_350 |
0.1041836119 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 10 |
Hb_002249_160 |
0.1042521535 |
- |
- |
- |
| 11 |
Hb_000375_030 |
0.1054732744 |
- |
- |
- |
| 12 |
Hb_004899_030 |
0.1055736953 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 13 |
Hb_028707_090 |
0.1090273964 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 14 |
Hb_009151_020 |
0.11014137 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 15 |
Hb_000640_020 |
0.1107035483 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 16 |
Hb_003436_050 |
0.1118493302 |
- |
- |
arf gtpase-activating protein, putative [Ricinus communis] |
| 17 |
Hb_017895_020 |
0.1122206469 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 18 |
Hb_002301_270 |
0.1123134898 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 19 |
Hb_001489_070 |
0.1126644462 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 20 |
Hb_001473_080 |
0.1141352439 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |