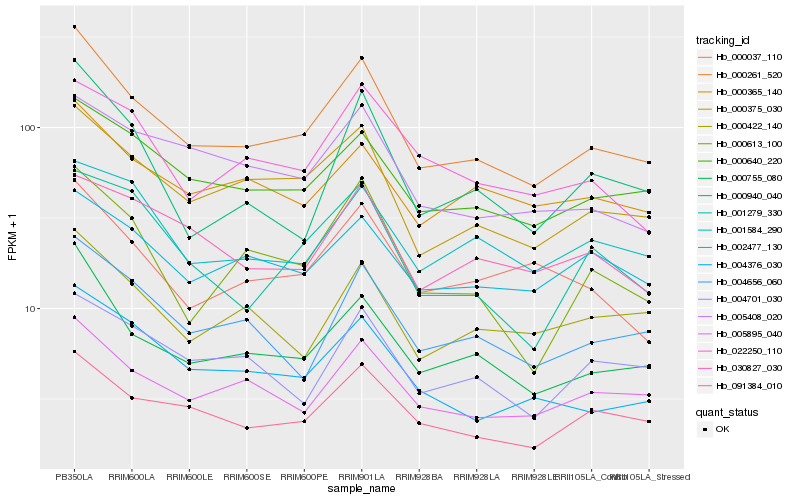
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_520 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633192 [Jatropha curcas] |
| 2 |
Hb_000755_080 |
0.0671140638 |
- |
- |
hypothetical protein RCOM_1016710 [Ricinus communis] |
| 3 |
Hb_000375_030 |
0.0757610945 |
- |
- |
- |
| 4 |
Hb_091384_010 |
0.0858548493 |
- |
- |
hypothetical protein JCGZ_09207 [Jatropha curcas] |
| 5 |
Hb_000640_220 |
0.0886596546 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 6 |
Hb_005895_040 |
0.0894277874 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 7 |
Hb_004376_030 |
0.0955737751 |
transcription factor |
TF Family: mTERF |
hypothetical protein PHAVU_005G060900g [Phaseolus vulgaris] |
| 8 |
Hb_004656_060 |
0.098633784 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 9 |
Hb_000613_100 |
0.1029010655 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 8 [Jatropha curcas] |
| 10 |
Hb_000365_140 |
0.1058258431 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase [Jatropha curcas] |
| 11 |
Hb_000037_110 |
0.108781548 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Populus euphratica] |
| 12 |
Hb_000422_140 |
0.1103198044 |
- |
- |
PREDICTED: apoptosis-inducing factor 2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000940_040 |
0.1107462477 |
- |
- |
PREDICTED: protein XAP5 CIRCADIAN TIMEKEEPER isoform X1 [Jatropha curcas] |
| 14 |
Hb_002477_130 |
0.1108372875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005408_020 |
0.1112154816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_022250_110 |
0.1115050876 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_001584_290 |
0.1118830756 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 18 |
Hb_001279_330 |
0.1151129142 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 19 |
Hb_030827_030 |
0.1151207759 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 20 |
Hb_004701_030 |
0.1151333967 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |