| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
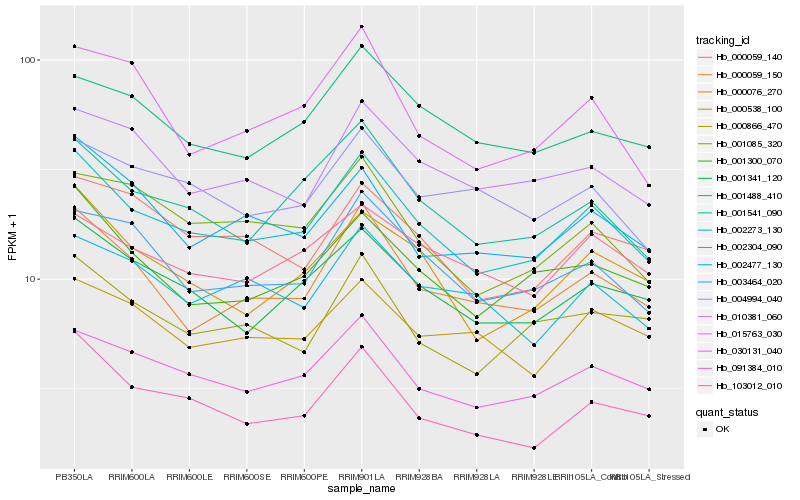
Hb_002273_130 |
0.0 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta [Gossypium raimondii] |
| 2 |
Hb_030131_040 |
0.0574114115 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 3 |
Hb_001341_120 |
0.0646616835 |
- |
- |
hypothetical protein B456_001G130300 [Gossypium raimondii] |
| 4 |
Hb_001541_090 |
0.0666567734 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000076_270 |
0.0696404464 |
- |
- |
PREDICTED: DDRGK domain-containing protein 1 [Populus euphratica] |
| 6 |
Hb_000059_150 |
0.0698822936 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein isoform 2 [Theobroma cacao] |
| 7 |
Hb_001300_070 |
0.0721493736 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 8 |
Hb_003464_020 |
0.0729520934 |
- |
- |
srpk, putative [Ricinus communis] |
| 9 |
Hb_001085_320 |
0.0780414281 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 10 |
Hb_010381_060 |
0.0815793641 |
- |
- |
DEAD-box ATP-dependent RNA helicase 20 -like protein [Gossypium arboreum] |
| 11 |
Hb_091384_010 |
0.08172966 |
- |
- |
hypothetical protein JCGZ_09207 [Jatropha curcas] |
| 12 |
Hb_000059_140 |
0.083402264 |
- |
- |
rnase h, putative [Ricinus communis] |
| 13 |
Hb_002477_130 |
0.0843286252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002304_090 |
0.0846072899 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX35 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000866_470 |
0.0847248006 |
- |
- |
PREDICTED: uncharacterized protein LOC105643015 [Jatropha curcas] |
| 16 |
Hb_103012_010 |
0.0850735625 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 17 |
Hb_001488_410 |
0.0853994995 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 18 |
Hb_015763_030 |
0.0864590446 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000538_100 |
0.0880948008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004994_040 |
0.0883717964 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |