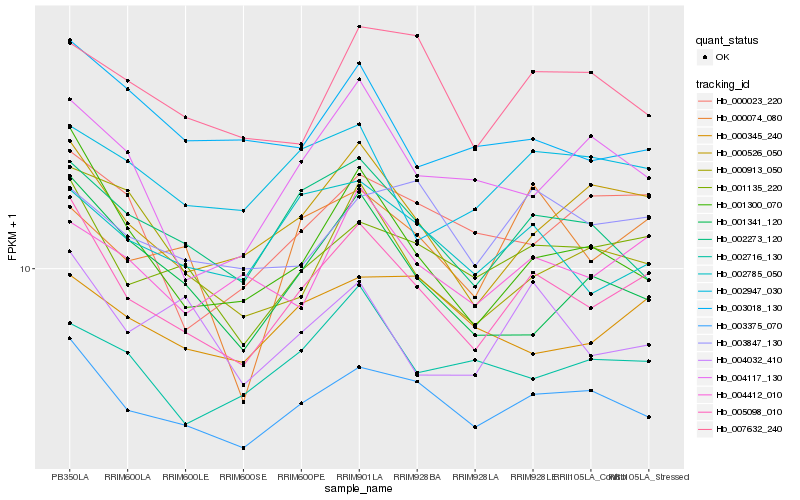
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005098_010 |
0.0 |
- |
- |
PREDICTED: L-aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase-like [Jatropha curcas] |
| 2 |
Hb_003375_070 |
0.0723684879 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 3 |
Hb_001135_220 |
0.0732668197 |
- |
- |
FGFR1 oncogene partner, putative [Ricinus communis] |
| 4 |
Hb_000526_050 |
0.0748746392 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001300_070 |
0.0794390564 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 6 |
Hb_002273_120 |
0.0904920427 |
- |
- |
PREDICTED: probable apyrase 6 [Jatropha curcas] |
| 7 |
Hb_002785_050 |
0.0957559699 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 8 |
Hb_004412_010 |
0.0976421965 |
- |
- |
- |
| 9 |
Hb_001341_120 |
0.099997081 |
- |
- |
hypothetical protein B456_001G130300 [Gossypium raimondii] |
| 10 |
Hb_000023_220 |
0.1027360698 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 18 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004032_410 |
0.103960753 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004117_130 |
0.105225892 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 13 |
Hb_000913_050 |
0.1053999415 |
- |
- |
hypothetical protein JCGZ_17329 [Jatropha curcas] |
| 14 |
Hb_002947_030 |
0.1057823227 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 15 |
Hb_003018_130 |
0.1061906245 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 16 |
Hb_002716_130 |
0.1064527376 |
- |
- |
PREDICTED: uncharacterized protein LOC105645470 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003847_130 |
0.1070113983 |
- |
- |
cytochrome C oxidase assembly protein cox11, putative [Ricinus communis] |
| 18 |
Hb_000345_240 |
0.10798652 |
- |
- |
PREDICTED: cyclin-H1-1 [Jatropha curcas] |
| 19 |
Hb_000074_080 |
0.1081332416 |
- |
- |
unknown [Populus trichocarpa] |
| 20 |
Hb_007632_240 |
0.1093774585 |
- |
- |
transporter, putative [Ricinus communis] |