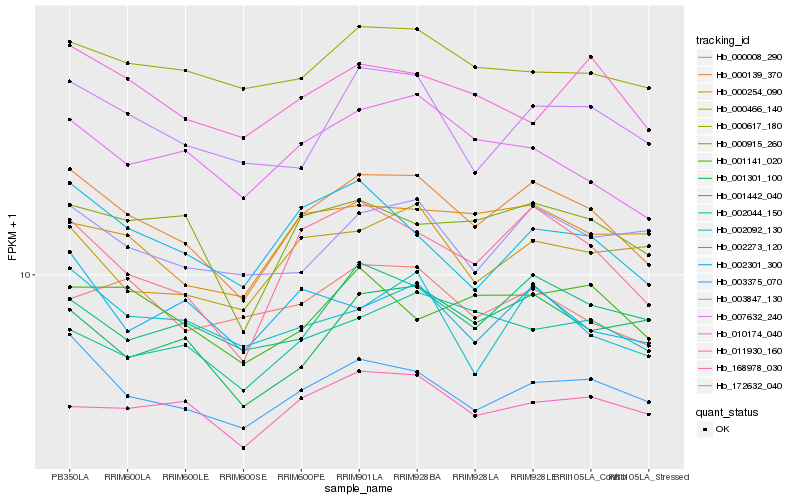
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_370 |
0.0 |
- |
- |
ubiquitin-activating enzyme E1c, putative [Ricinus communis] |
| 2 |
Hb_172632_040 |
0.056456631 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 isoform X1 [Amborella trichopoda] |
| 3 |
Hb_003375_070 |
0.0653897181 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 4 |
Hb_000915_260 |
0.0776442865 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |
| 5 |
Hb_010174_040 |
0.0806781426 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 6 |
Hb_007632_240 |
0.0813031553 |
- |
- |
transporter, putative [Ricinus communis] |
| 7 |
Hb_002273_120 |
0.0850602851 |
- |
- |
PREDICTED: probable apyrase 6 [Jatropha curcas] |
| 8 |
Hb_000254_090 |
0.0851177008 |
- |
- |
PREDICTED: la protein 2 [Jatropha curcas] |
| 9 |
Hb_001442_040 |
0.0856482563 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10 isoform X1 [Jatropha curcas] |
| 10 |
Hb_168978_030 |
0.086446569 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 8 [Jatropha curcas] |
| 11 |
Hb_003847_130 |
0.0870154825 |
- |
- |
cytochrome C oxidase assembly protein cox11, putative [Ricinus communis] |
| 12 |
Hb_002092_130 |
0.0879160274 |
- |
- |
PREDICTED: protein CASP-like [Oryza brachyantha] |
| 13 |
Hb_000617_180 |
0.0884957495 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 14 |
Hb_000466_140 |
0.0888563518 |
- |
- |
hypothetical protein JCGZ_02122 [Jatropha curcas] |
| 15 |
Hb_001141_020 |
0.0899636215 |
- |
- |
PREDICTED: stress-induced-phosphoprotein 1 [Jatropha curcas] |
| 16 |
Hb_002044_150 |
0.0907446154 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 17 |
Hb_002301_300 |
0.0917750085 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 18 |
Hb_001301_100 |
0.0923003452 |
- |
- |
transporter, putative [Ricinus communis] |
| 19 |
Hb_000008_290 |
0.0923384335 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 20 |
Hb_011930_160 |
0.0925960613 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 38-like [Jatropha curcas] |