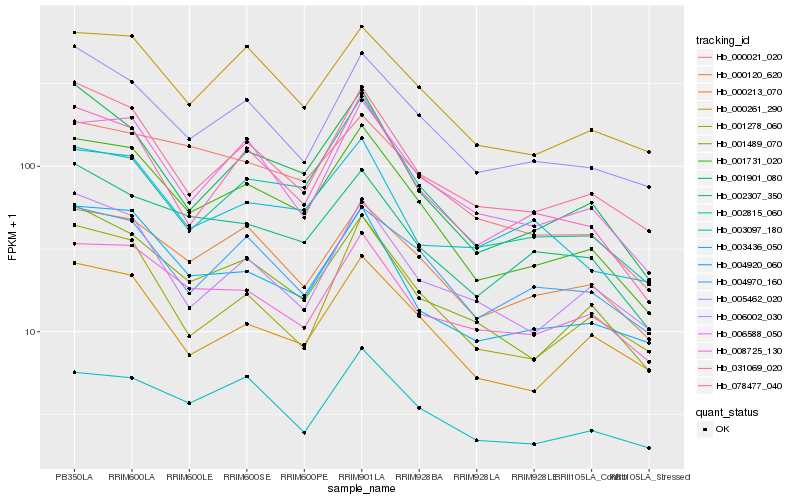
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001731_020 |
0.0 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 2 |
Hb_031069_020 |
0.0788440922 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 3-like [Jatropha curcas] |
| 3 |
Hb_001489_070 |
0.0804867032 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 4 |
Hb_078477_040 |
0.0847648348 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 5 |
Hb_004920_060 |
0.0854914333 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 6 |
Hb_000261_290 |
0.0857403431 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 7 |
Hb_006588_050 |
0.0861813705 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 8 |
Hb_001901_080 |
0.0886846645 |
- |
- |
PREDICTED: xylose isomerase [Jatropha curcas] |
| 9 |
Hb_005462_020 |
0.0903044356 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 10 |
Hb_000120_620 |
0.0933298911 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 11 |
Hb_000213_070 |
0.094920041 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor At2g41710 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000021_020 |
0.1008196829 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Malus domestica] |
| 13 |
Hb_002307_350 |
0.1013582743 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 14 |
Hb_003097_180 |
0.1029308731 |
- |
- |
PREDICTED: crossover junction endonuclease MUS81 [Jatropha curcas] |
| 15 |
Hb_008725_130 |
0.1031466885 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 16 |
Hb_004970_160 |
0.1039406724 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_002815_060 |
0.1054901742 |
- |
- |
hypothetical protein B456_002G110500 [Gossypium raimondii] |
| 18 |
Hb_001278_060 |
0.1058217884 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 19 |
Hb_006002_030 |
0.1087392103 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003436_050 |
0.1107175646 |
- |
- |
arf gtpase-activating protein, putative [Ricinus communis] |