| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
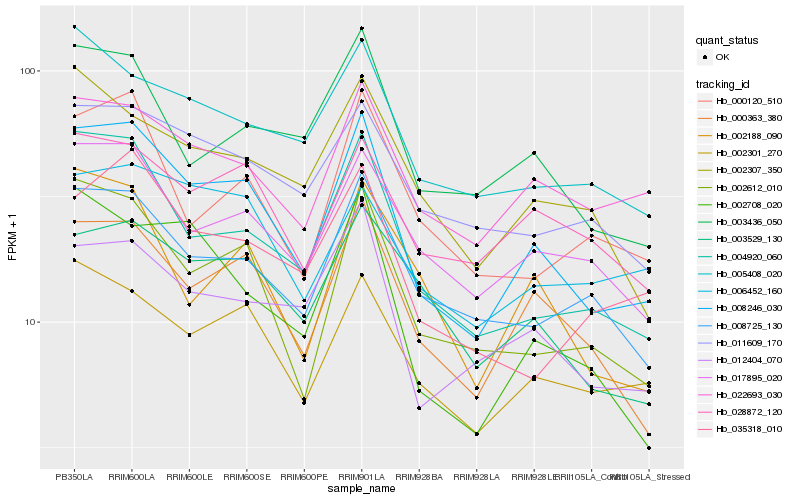
Hb_008246_030 |
0.0 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 2 |
Hb_000363_380 |
0.0847008134 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |
| 3 |
Hb_004920_060 |
0.0884856332 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 4 |
Hb_002612_010 |
0.0951928715 |
- |
- |
hypothetical protein JCGZ_19269 [Jatropha curcas] |
| 5 |
Hb_002301_270 |
0.0984827716 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 6 |
Hb_012404_070 |
0.1050346707 |
- |
- |
PREDICTED: uncharacterized protein LOC105629465 [Jatropha curcas] |
| 7 |
Hb_017895_020 |
0.1080201752 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 8 |
Hb_008725_130 |
0.1105560828 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 9 |
Hb_003436_050 |
0.1132854597 |
- |
- |
arf gtpase-activating protein, putative [Ricinus communis] |
| 10 |
Hb_005408_020 |
0.1142336098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003529_130 |
0.1142681321 |
- |
- |
PREDICTED: ribosome biogenesis protein BOP1 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_002708_020 |
0.1144246396 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: uncharacterized protein At5g08430-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_022693_030 |
0.1163912074 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 14 |
Hb_028872_120 |
0.1166810201 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 15 |
Hb_002188_090 |
0.1195466892 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X1 [Jatropha curcas] |
| 16 |
Hb_002307_350 |
0.1196058934 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 17 |
Hb_011609_170 |
0.1200804564 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 18 |
Hb_000120_510 |
0.1205560439 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_035318_010 |
0.1208954185 |
- |
- |
PREDICTED: uncharacterized protein LOC105645250 [Jatropha curcas] |
| 20 |
Hb_006452_160 |
0.1237655583 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |