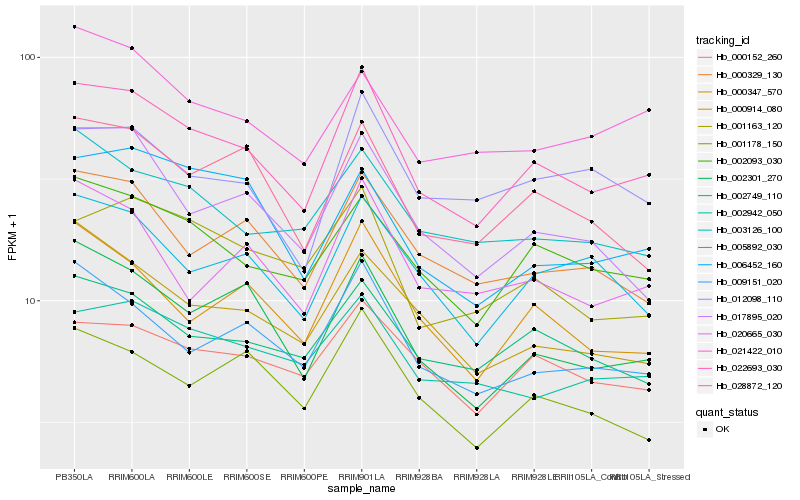
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022693_030 |
0.0 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 2 |
Hb_002093_030 |
0.069637709 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 3 |
Hb_002301_270 |
0.0785183555 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 4 |
Hb_009151_020 |
0.0792220503 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 5 |
Hb_002749_110 |
0.0815106013 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005892_030 |
0.0836876763 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 7 |
Hb_000347_570 |
0.0836963873 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 8 |
Hb_000329_130 |
0.0843157926 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 9 |
Hb_028872_120 |
0.0845886845 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 10 |
Hb_000152_260 |
0.0845941081 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 11 |
Hb_002942_050 |
0.0870936302 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 12 |
Hb_003126_100 |
0.090001273 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 13 |
Hb_017895_020 |
0.090234597 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 14 |
Hb_000914_080 |
0.0905560583 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_020665_030 |
0.0906250603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001163_120 |
0.0909065721 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_001178_150 |
0.0911017485 |
- |
- |
PREDICTED: uncharacterized protein LOC105635484 [Jatropha curcas] |
| 18 |
Hb_012098_110 |
0.0930014274 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 19 |
Hb_021422_010 |
0.0947471137 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 20 |
Hb_006452_160 |
0.095653468 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |