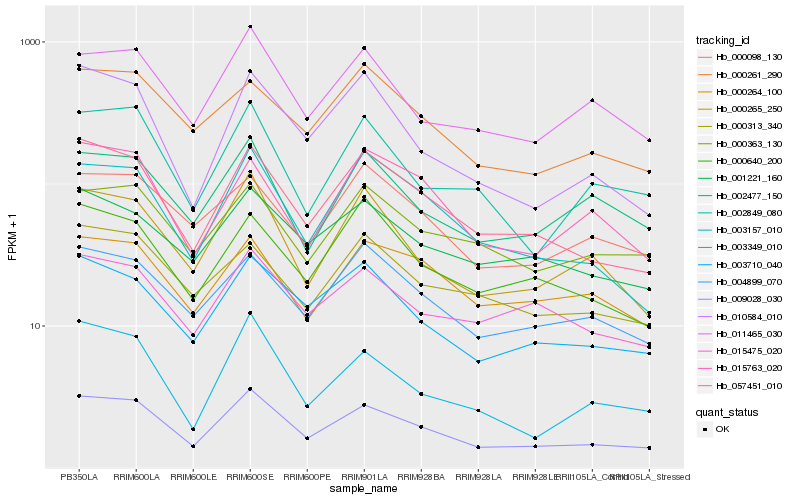
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009028_030 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
NBS type disease resistance protein, putative [Theobroma cacao] |
| 2 |
Hb_057451_010 |
0.0648394793 |
- |
- |
PREDICTED: uncharacterized protein LOC105635588 [Jatropha curcas] |
| 3 |
Hb_003710_040 |
0.0820364796 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 4 |
Hb_000265_250 |
0.090371609 |
- |
- |
auxilin, putative [Ricinus communis] |
| 5 |
Hb_003157_010 |
0.0914889087 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 6 |
Hb_004899_070 |
0.0929797071 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 7 |
Hb_011465_030 |
0.0933825527 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 8 |
Hb_010584_010 |
0.0977854231 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 9 |
Hb_001221_160 |
0.1007097372 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 10 |
Hb_002849_080 |
0.1007818336 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 11 |
Hb_015475_020 |
0.1010408739 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 12 |
Hb_003349_010 |
0.1023722932 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CICLE_v10023911mg [Citrus clementina] |
| 13 |
Hb_000640_200 |
0.1030374188 |
- |
- |
hypothetical protein RCOM_1346600 [Ricinus communis] |
| 14 |
Hb_000363_130 |
0.1037502328 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000261_290 |
0.103898958 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 16 |
Hb_015763_020 |
0.1046763976 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 17 |
Hb_000313_340 |
0.1059502754 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 18 |
Hb_000264_100 |
0.1062492201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002477_150 |
0.1070002356 |
transcription factor |
TF Family: Alfin-like |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_000098_130 |
0.1078634737 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |