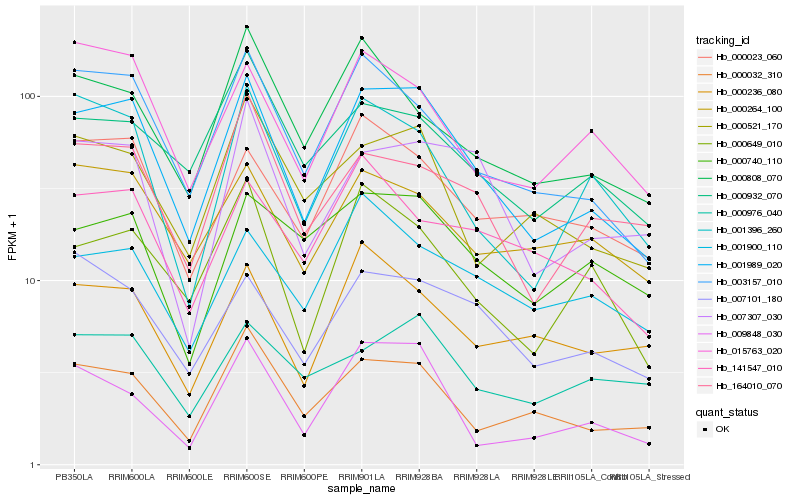
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001989_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_15521 [Jatropha curcas] |
| 2 |
Hb_003157_010 |
0.1103100255 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 3 |
Hb_000032_310 |
0.1267550186 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 4 |
Hb_000649_010 |
0.132169286 |
- |
- |
PREDICTED: adagio protein 3 [Jatropha curcas] |
| 5 |
Hb_000023_060 |
0.1329126943 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 6 |
Hb_001396_260 |
0.1329566567 |
- |
- |
PREDICTED: F-box only protein 6 [Jatropha curcas] |
| 7 |
Hb_007101_180 |
0.1340533449 |
- |
- |
PREDICTED: DNA polymerase beta [Jatropha curcas] |
| 8 |
Hb_007307_030 |
0.1374043994 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000521_170 |
0.1391347911 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |
| 10 |
Hb_000264_100 |
0.1402292725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_015763_020 |
0.1406031848 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 12 |
Hb_164010_070 |
0.1417559744 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 13 |
Hb_000236_080 |
0.146340161 |
- |
- |
PREDICTED: small subunit processome component 20 homolog [Jatropha curcas] |
| 14 |
Hb_000808_070 |
0.1470704211 |
- |
- |
PREDICTED: importin subunit alpha-2 [Jatropha curcas] |
| 15 |
Hb_000932_070 |
0.1479955502 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 16 |
Hb_009848_030 |
0.1483336417 |
- |
- |
long-chain-fatty-acid CoA ligase, putative [Ricinus communis] |
| 17 |
Hb_000740_110 |
0.1486637489 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01850 [Jatropha curcas] |
| 18 |
Hb_141547_010 |
0.1487669497 |
- |
- |
PREDICTED: ARF guanine-nucleotide exchange factor GNOM [Jatropha curcas] |
| 19 |
Hb_001900_110 |
0.1493900499 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 20 |
Hb_000976_040 |
0.1526877087 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |