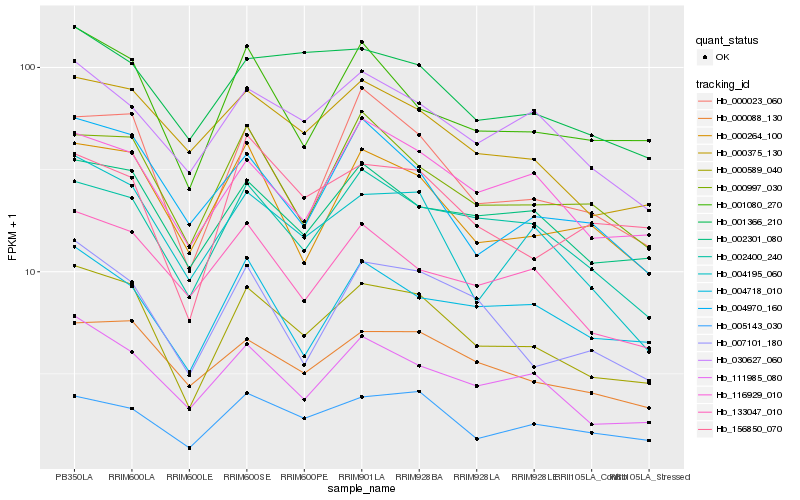
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000589_040 |
0.0 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At1g12280 [Jatropha curcas] |
| 2 |
Hb_111985_080 |
0.0898524275 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g19720 [Jatropha curcas] |
| 3 |
Hb_000023_060 |
0.0947707554 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 4 |
Hb_002301_080 |
0.0963151703 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000088_130 |
0.0971117524 |
- |
- |
hypothetical protein JCGZ_02447 [Jatropha curcas] |
| 6 |
Hb_002400_240 |
0.09721922 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 7 |
Hb_005143_030 |
0.09824782 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 8 |
Hb_001080_270 |
0.0985295951 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 9 |
Hb_000375_130 |
0.1009304819 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_116929_010 |
0.1020452087 |
- |
- |
PREDICTED: nuclear-pore anchor isoform X2 [Jatropha curcas] |
| 11 |
Hb_000264_100 |
0.1021458362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004195_060 |
0.1023403433 |
- |
- |
PREDICTED: uncharacterized protein LOC105803780 isoform X1 [Gossypium raimondii] |
| 13 |
Hb_000997_030 |
0.103003435 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 14 |
Hb_030627_060 |
0.1047337166 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 15 |
Hb_007101_180 |
0.1067067184 |
- |
- |
PREDICTED: DNA polymerase beta [Jatropha curcas] |
| 16 |
Hb_001366_210 |
0.1094471408 |
- |
- |
cathepsin B, putative [Ricinus communis] |
| 17 |
Hb_004718_010 |
0.1098087802 |
- |
- |
PREDICTED: uncharacterized protein LOC105638290 [Jatropha curcas] |
| 18 |
Hb_133047_010 |
0.1100839693 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_156850_070 |
0.1106356526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004970_160 |
0.1129179191 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |