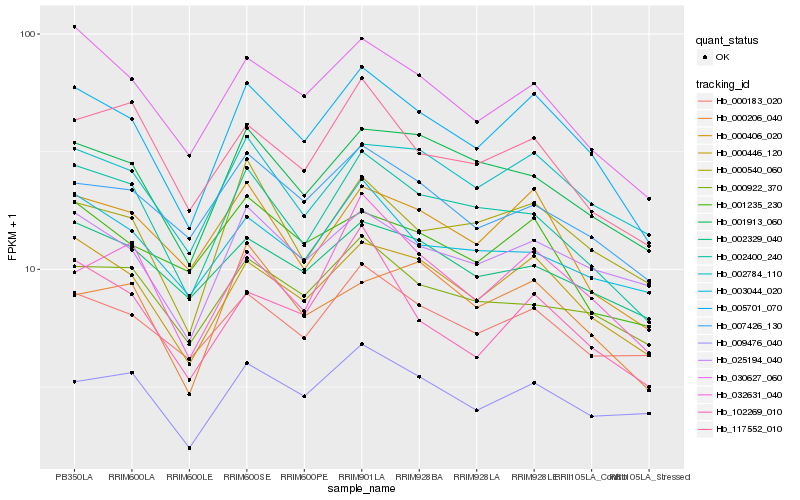
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005701_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized TPR repeat-containing protein At1g05150-like [Jatropha curcas] |
| 2 |
Hb_000446_120 |
0.0531952519 |
- |
- |
PREDICTED: uncharacterized transmembrane protein DDB_G0289901 [Jatropha curcas] |
| 3 |
Hb_030627_060 |
0.0747245268 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 4 |
Hb_002400_240 |
0.0762471382 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 5 |
Hb_002784_110 |
0.0768790662 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 6 |
Hb_000406_020 |
0.0800562086 |
- |
- |
- |
| 7 |
Hb_025194_040 |
0.0822542213 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000540_060 |
0.0836313775 |
- |
- |
PREDICTED: uncharacterized protein LOC105628177 [Jatropha curcas] |
| 9 |
Hb_001913_060 |
0.0855554124 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 10 |
Hb_007426_130 |
0.0887593331 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 11 |
Hb_000183_020 |
0.0893202865 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 12 |
Hb_032631_040 |
0.0918234728 |
- |
- |
PREDICTED: uncharacterized protein LOC105638769 [Jatropha curcas] |
| 13 |
Hb_009476_040 |
0.0921763129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000922_370 |
0.0928474014 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 15 |
Hb_117552_010 |
0.0943041017 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_102269_010 |
0.0959505174 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |
| 17 |
Hb_002329_040 |
0.0961670768 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 18 |
Hb_001235_230 |
0.0965147779 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 19 |
Hb_000206_040 |
0.0970862381 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 20 |
Hb_003044_020 |
0.0974259451 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |