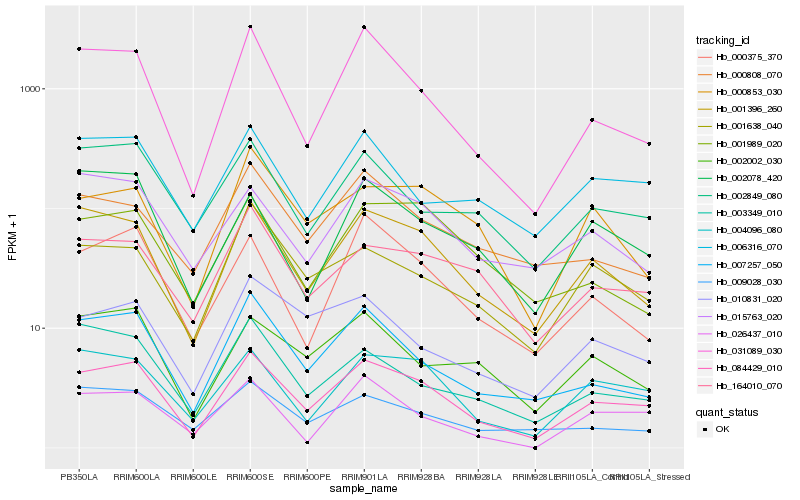
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_084429_010 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Populus euphratica] |
| 2 |
Hb_001396_260 |
0.0953868636 |
- |
- |
PREDICTED: F-box only protein 6 [Jatropha curcas] |
| 3 |
Hb_004096_080 |
0.1170239866 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 4 |
Hb_031089_030 |
0.117175815 |
- |
- |
ubiquitin-conjugating enzyme E2 28 [Arabidopsis thaliana] |
| 5 |
Hb_007257_050 |
0.1258827559 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_000375_370 |
0.1398757048 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_002849_080 |
0.1406148029 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 8 |
Hb_164010_070 |
0.1408060117 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 9 |
Hb_006316_070 |
0.1427189186 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 10 |
Hb_002002_030 |
0.1434616959 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 11 |
Hb_009028_030 |
0.1454799464 |
desease resistance |
Gene Name: NB-ARC |
NBS type disease resistance protein, putative [Theobroma cacao] |
| 12 |
Hb_001638_040 |
0.1455751533 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 13 |
Hb_015763_020 |
0.1458683212 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 14 |
Hb_010831_020 |
0.1472723267 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 15 |
Hb_003349_010 |
0.1532608833 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CICLE_v10023911mg [Citrus clementina] |
| 16 |
Hb_000853_030 |
0.1544111811 |
- |
- |
sucrose synthase 2 [Hevea brasiliensis] |
| 17 |
Hb_002078_420 |
0.1546614667 |
- |
- |
PREDICTED: F-box only protein 6 [Jatropha curcas] |
| 18 |
Hb_001989_020 |
0.1572577619 |
- |
- |
hypothetical protein JCGZ_15521 [Jatropha curcas] |
| 19 |
Hb_000808_070 |
0.1576042874 |
- |
- |
PREDICTED: importin subunit alpha-2 [Jatropha curcas] |
| 20 |
Hb_026437_010 |
0.1578691063 |
- |
- |
- |