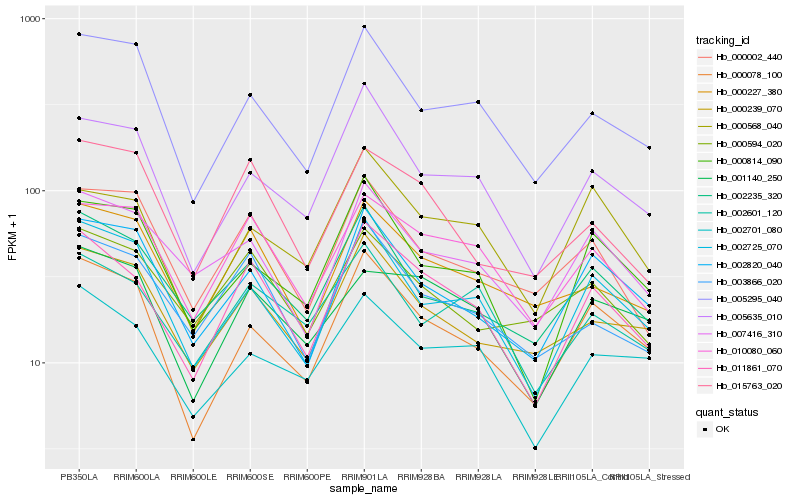
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011861_070 |
0.0 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_010080_060 |
0.0964167094 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 3 |
Hb_003866_020 |
0.1071812551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000078_100 |
0.1099512152 |
- |
- |
PREDICTED: topless-related protein 4-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_005635_010 |
0.1117384277 |
- |
- |
PREDICTED: cleft lip and palate transmembrane protein 1 homolog [Jatropha curcas] |
| 6 |
Hb_002725_070 |
0.1118208968 |
- |
- |
hypothetical protein PRUPE_ppa005949mg [Prunus persica] |
| 7 |
Hb_000227_380 |
0.1126885892 |
- |
- |
PREDICTED: protein TIME FOR COFFEE isoform X3 [Jatropha curcas] |
| 8 |
Hb_007416_310 |
0.1155470577 |
- |
- |
PREDICTED: zinc finger protein 622 [Jatropha curcas] |
| 9 |
Hb_002235_320 |
0.1165377945 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000239_070 |
0.1181148378 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002820_040 |
0.1204388338 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002701_080 |
0.1215480502 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like [Jatropha curcas] |
| 13 |
Hb_015763_020 |
0.1217273409 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 14 |
Hb_002601_120 |
0.1227037873 |
- |
- |
PREDICTED: uncharacterized protein LOC105634921 [Jatropha curcas] |
| 15 |
Hb_001140_250 |
0.1228890781 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_000002_440 |
0.123174745 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 17 |
Hb_005295_040 |
0.1236570325 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP47-like [Jatropha curcas] |
| 18 |
Hb_000814_090 |
0.1258856311 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000568_040 |
0.1263403362 |
- |
- |
PREDICTED: mRNA-decapping enzyme subunit 2 [Jatropha curcas] |
| 20 |
Hb_000594_020 |
0.1271693767 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |