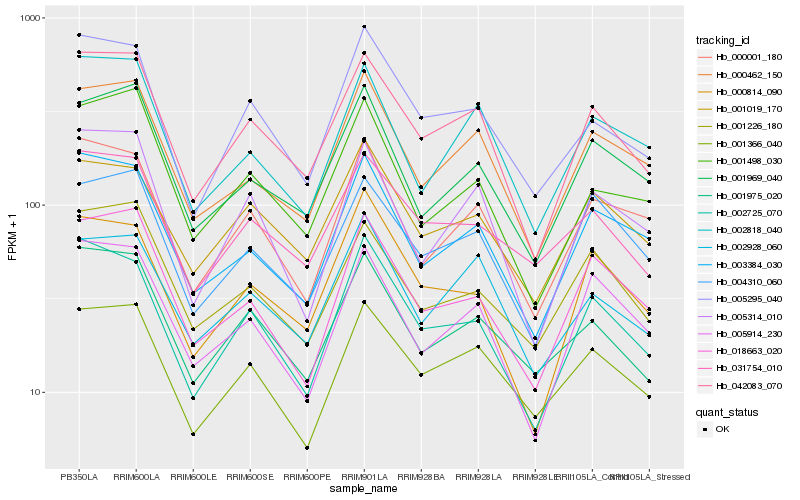
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_042083_070 |
0.0 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 2 |
Hb_002725_070 |
0.0649262151 |
- |
- |
hypothetical protein PRUPE_ppa005949mg [Prunus persica] |
| 3 |
Hb_005295_040 |
0.0762936634 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP47-like [Jatropha curcas] |
| 4 |
Hb_000462_150 |
0.0774262462 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Jatropha curcas] |
| 5 |
Hb_005914_230 |
0.0781657519 |
- |
- |
PREDICTED: pre-mRNA-processing factor 19 homolog 1 [Jatropha curcas] |
| 6 |
Hb_003384_030 |
0.0783933019 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit beta-like [Jatropha curcas] |
| 7 |
Hb_004310_060 |
0.079097308 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_001975_020 |
0.0796156365 |
- |
- |
PREDICTED: uncharacterized protein LOC105637186 [Jatropha curcas] |
| 9 |
Hb_001019_170 |
0.0813723442 |
- |
- |
PREDICTED: uncharacterized protein LOC105639322 [Jatropha curcas] |
| 10 |
Hb_001366_040 |
0.0820138104 |
transcription factor |
TF Family: SNF2 |
PREDICTED: endoribonuclease Dicer homolog 1 [Jatropha curcas] |
| 11 |
Hb_001969_040 |
0.0834197368 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 12 |
Hb_002928_060 |
0.0834779887 |
- |
- |
PREDICTED: pre-rRNA-processing protein TSR1 homolog [Jatropha curcas] |
| 13 |
Hb_005314_010 |
0.0844741465 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 14 |
Hb_018663_020 |
0.084521377 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000814_090 |
0.0850710126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001226_180 |
0.085765101 |
- |
- |
peptidyl-prolyl cis-trans isomerase, putative [Ricinus communis] |
| 17 |
Hb_000001_180 |
0.0862607868 |
- |
- |
PREDICTED: uncharacterized protein LOC105648978 [Jatropha curcas] |
| 18 |
Hb_031754_010 |
0.0865809426 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_001498_030 |
0.0871300303 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit E [Populus euphratica] |
| 20 |
Hb_002818_040 |
0.087375711 |
- |
- |
tRNA synthetase-related family protein [Populus trichocarpa] |