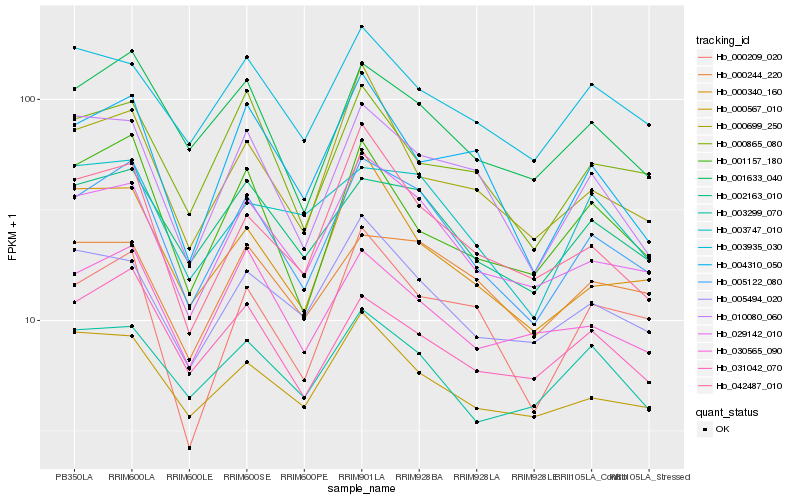
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005122_080 |
0.0 |
- |
- |
prokaryotic DNA topoisomerase, putative [Ricinus communis] |
| 2 |
Hb_029142_010 |
0.0637566761 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 3 |
Hb_002163_010 |
0.0785046557 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 4 |
Hb_001633_040 |
0.0816642185 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 5 |
Hb_010080_060 |
0.0892335474 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_031042_070 |
0.0953899812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005494_020 |
0.0968017372 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 8 |
Hb_000567_010 |
0.0986089796 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 9 |
Hb_000244_220 |
0.099329017 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000865_080 |
0.0998096319 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 11 |
Hb_030565_090 |
0.0998760682 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |
| 12 |
Hb_042487_010 |
0.1006240913 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 9-like [Jatropha curcas] |
| 13 |
Hb_001157_180 |
0.1012930179 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_003935_030 |
0.1022980896 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 15 |
Hb_000340_160 |
0.1024650394 |
- |
- |
PREDICTED: elongation factor Tu, mitochondrial [Jatropha curcas] |
| 16 |
Hb_003299_070 |
0.1029006733 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 17 |
Hb_003747_010 |
0.1031331449 |
- |
- |
hypothetical protein JCGZ_24080 [Jatropha curcas] |
| 18 |
Hb_000209_020 |
0.1048743297 |
- |
- |
PREDICTED: uncharacterized protein At4g26450 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000699_250 |
0.1052381621 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 20 |
Hb_004310_050 |
0.1052438611 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |