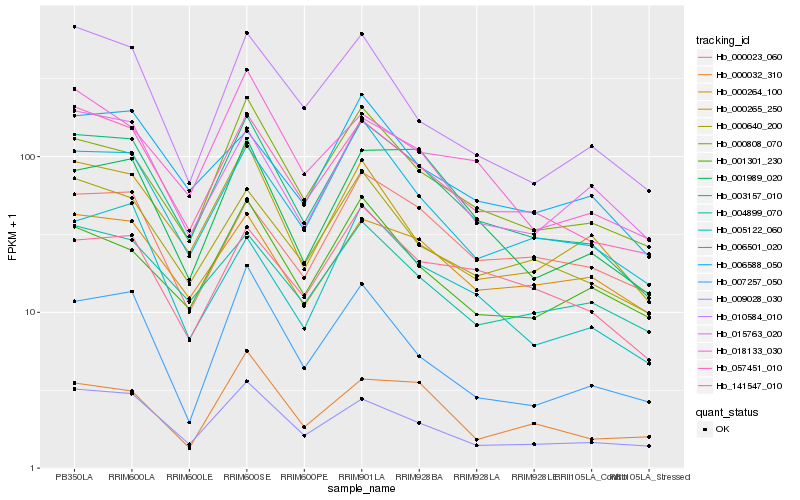
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003157_010 |
0.0 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 2 |
Hb_057451_010 |
0.0727072798 |
- |
- |
PREDICTED: uncharacterized protein LOC105635588 [Jatropha curcas] |
| 3 |
Hb_006501_020 |
0.0846301457 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 4 |
Hb_000808_070 |
0.0877392084 |
- |
- |
PREDICTED: importin subunit alpha-2 [Jatropha curcas] |
| 5 |
Hb_009028_030 |
0.0914889087 |
desease resistance |
Gene Name: NB-ARC |
NBS type disease resistance protein, putative [Theobroma cacao] |
| 6 |
Hb_000640_200 |
0.0970953346 |
- |
- |
hypothetical protein RCOM_1346600 [Ricinus communis] |
| 7 |
Hb_001989_020 |
0.1103100255 |
- |
- |
hypothetical protein JCGZ_15521 [Jatropha curcas] |
| 8 |
Hb_004899_070 |
0.111232095 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 9 |
Hb_015763_020 |
0.1116403279 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 10 |
Hb_001301_230 |
0.11178096 |
- |
- |
PREDICTED: uncharacterized protein LOC105632949 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000265_250 |
0.1124216751 |
- |
- |
auxilin, putative [Ricinus communis] |
| 12 |
Hb_141547_010 |
0.1136522647 |
- |
- |
PREDICTED: ARF guanine-nucleotide exchange factor GNOM [Jatropha curcas] |
| 13 |
Hb_006588_050 |
0.1153542978 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 14 |
Hb_010584_010 |
0.1154109982 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 15 |
Hb_007257_050 |
0.1170397509 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_018133_030 |
0.1191507719 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 17 |
Hb_000023_060 |
0.1191974297 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 18 |
Hb_000264_100 |
0.1194925075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005122_060 |
0.1203111151 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_000032_310 |
0.121322262 |
- |
- |
ring finger protein, putative [Ricinus communis] |