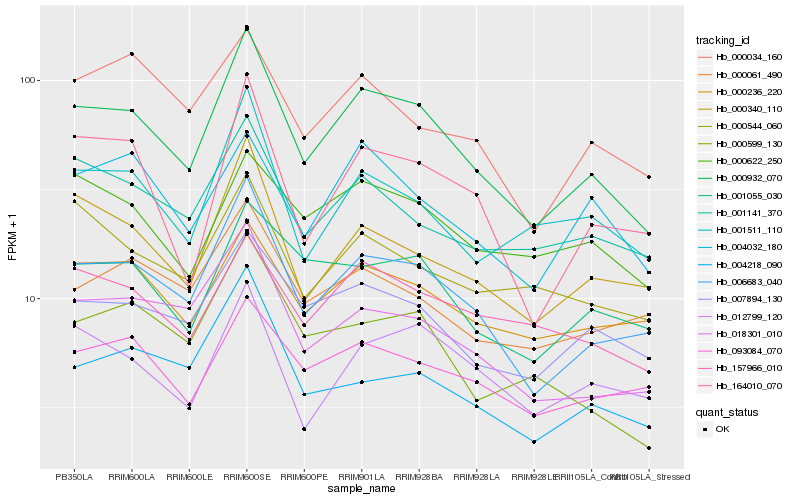
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000932_070 |
0.0 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 2 |
Hb_006683_040 |
0.0655503548 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 3 |
Hb_007894_130 |
0.0983963182 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 4 |
Hb_000340_110 |
0.0987393891 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 5 |
Hb_018301_010 |
0.0987862286 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 6 |
Hb_001511_110 |
0.1051926125 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_157966_010 |
0.1072690822 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 8 |
Hb_000236_220 |
0.1075222246 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 9 |
Hb_164010_070 |
0.1075871768 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 10 |
Hb_001055_030 |
0.1079963789 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001141_370 |
0.1082086733 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 12 |
Hb_093084_070 |
0.1117029212 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 13 |
Hb_000599_130 |
0.1131218867 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 14 |
Hb_012799_120 |
0.1131861862 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004032_180 |
0.1139928632 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 16 |
Hb_000622_250 |
0.1142926215 |
- |
- |
atpob1, putative [Ricinus communis] |
| 17 |
Hb_000034_160 |
0.11600986 |
- |
- |
hypothetical protein EUGRSUZ_F02151 [Eucalyptus grandis] |
| 18 |
Hb_000544_060 |
0.1160576224 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 19 |
Hb_004218_090 |
0.1172724144 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000061_490 |
0.1179144482 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |