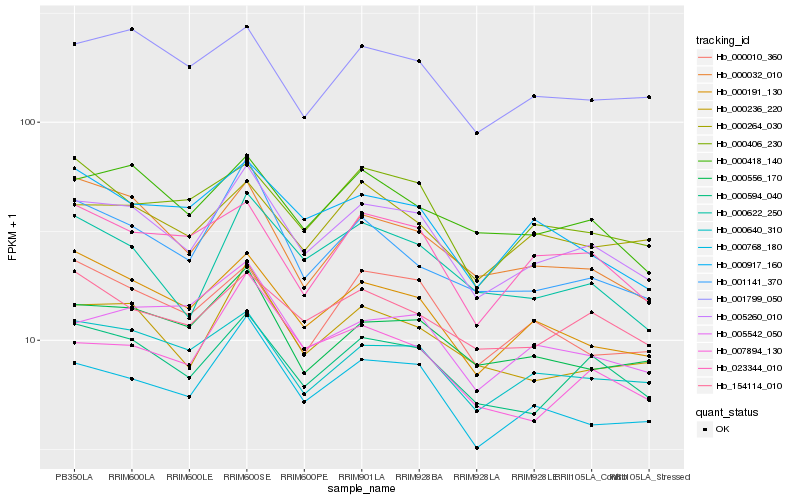
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005260_010 |
0.0 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000594_040 |
0.0554159284 |
- |
- |
hypothetical protein L484_013922 [Morus notabilis] |
| 3 |
Hb_000768_180 |
0.0565641351 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 4 |
Hb_000236_220 |
0.0634552197 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 5 |
Hb_000640_310 |
0.0683627061 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 6 |
Hb_000191_130 |
0.06973725 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 7 |
Hb_000418_140 |
0.0705136609 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 8 |
Hb_000556_170 |
0.0710959465 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001799_050 |
0.07396625 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 10 |
Hb_023344_010 |
0.0768404029 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000622_250 |
0.0768920947 |
- |
- |
atpob1, putative [Ricinus communis] |
| 12 |
Hb_154114_010 |
0.0773891348 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 13 |
Hb_000264_030 |
0.078302284 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 14 |
Hb_000406_230 |
0.0793576273 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 15 |
Hb_000032_010 |
0.0805126933 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 16 |
Hb_000010_360 |
0.0820222258 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 17 |
Hb_000917_160 |
0.0827090872 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001141_370 |
0.0828816168 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 19 |
Hb_005542_050 |
0.0829049107 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 20 |
Hb_007894_130 |
0.0837844188 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |