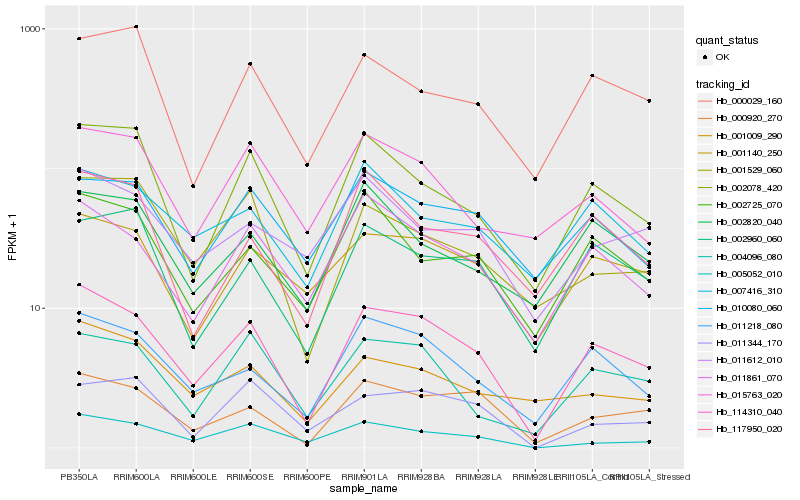
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114310_040 |
0.0 |
- |
- |
PREDICTED: purple acid phosphatase 2-like [Jatropha curcas] |
| 2 |
Hb_011218_080 |
0.1196330847 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 3 |
Hb_000920_270 |
0.1299530096 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |
| 4 |
Hb_002078_420 |
0.1318032918 |
- |
- |
PREDICTED: F-box only protein 6 [Jatropha curcas] |
| 5 |
Hb_011861_070 |
0.1344691798 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 6 |
Hb_004096_080 |
0.1425842281 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 7 |
Hb_005052_010 |
0.1472330411 |
- |
- |
PREDICTED: uncharacterized protein LOC103339315 [Prunus mume] |
| 8 |
Hb_001140_250 |
0.14763415 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_002960_060 |
0.1544983208 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 28 [Jatropha curcas] |
| 10 |
Hb_002725_070 |
0.1545088895 |
- |
- |
hypothetical protein PRUPE_ppa005949mg [Prunus persica] |
| 11 |
Hb_015763_020 |
0.1617585115 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 12 |
Hb_010080_060 |
0.1625021498 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 13 |
Hb_001529_060 |
0.1625699381 |
- |
- |
PREDICTED: U4/U6.U5 small nuclear ribonucleoprotein 27 kDa protein [Jatropha curcas] |
| 14 |
Hb_000029_160 |
0.162906811 |
- |
- |
Nitrilase, putative [Ricinus communis] |
| 15 |
Hb_007416_310 |
0.1635405603 |
- |
- |
PREDICTED: zinc finger protein 622 [Jatropha curcas] |
| 16 |
Hb_117950_020 |
0.1639916865 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Jatropha curcas] |
| 17 |
Hb_001009_290 |
0.1642975445 |
- |
- |
PREDICTED: elongator complex protein 3-like [Populus euphratica] |
| 18 |
Hb_002820_040 |
0.1646387044 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011344_170 |
0.1658151633 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 4-like, partial [Camelina sativa] |
| 20 |
Hb_011612_010 |
0.1699176919 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |