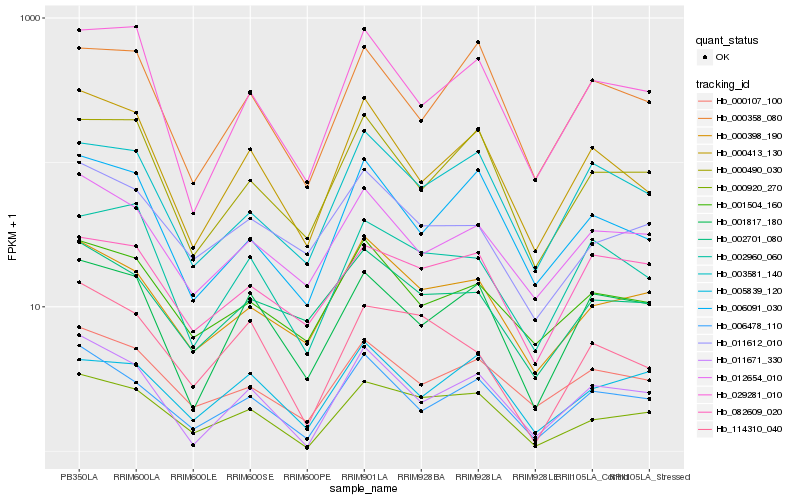
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000920_270 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |
| 2 |
Hb_000398_190 |
0.1273426504 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 3 |
Hb_114310_040 |
0.1299530096 |
- |
- |
PREDICTED: purple acid phosphatase 2-like [Jatropha curcas] |
| 4 |
Hb_006478_110 |
0.1335338119 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 5 |
Hb_000490_030 |
0.1390080085 |
- |
- |
DNA-directed RNA polymerase II subunit, putative [Ricinus communis] |
| 6 |
Hb_011671_330 |
0.140967886 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 4 [Jatropha curcas] |
| 7 |
Hb_006091_030 |
0.143792104 |
desease resistance |
Gene Name: AAA |
PREDICTED: cell division control protein 48 homolog C-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000107_100 |
0.1451461863 |
- |
- |
hypothetical protein JCGZ_16208 [Jatropha curcas] |
| 9 |
Hb_005839_120 |
0.1453603053 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 10 |
Hb_029281_010 |
0.1456515291 |
- |
- |
hypothetical protein JCGZ_04025 [Jatropha curcas] |
| 11 |
Hb_000413_130 |
0.1461199869 |
- |
- |
PREDICTED: uncharacterized protein LOC105649409 [Jatropha curcas] |
| 12 |
Hb_001817_180 |
0.1492473106 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 13 |
Hb_003581_140 |
0.1493637142 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1-like [Nelumbo nucifera] |
| 14 |
Hb_002960_060 |
0.1507149923 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 28 [Jatropha curcas] |
| 15 |
Hb_000358_080 |
0.1546574632 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 16 |
Hb_001504_160 |
0.1586258704 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 17 |
Hb_011612_010 |
0.1586413097 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 18 |
Hb_082609_020 |
0.1587050209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002701_080 |
0.1590257027 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like [Jatropha curcas] |
| 20 |
Hb_012654_010 |
0.1602925604 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |