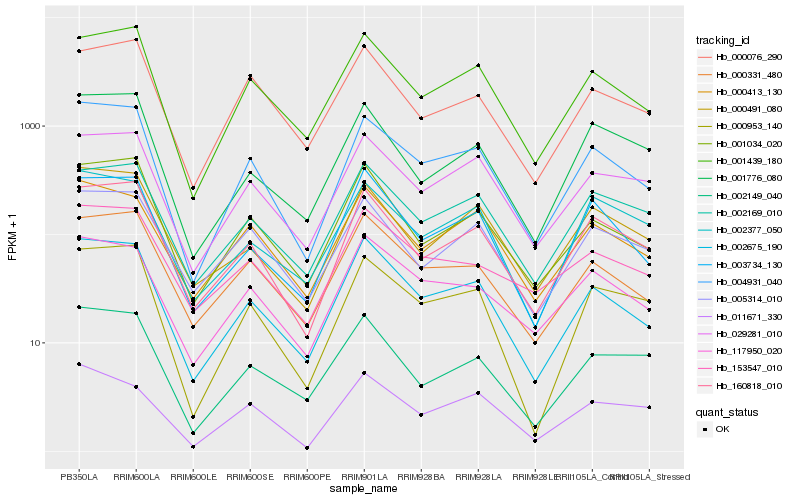
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004931_040 |
0.0 |
- |
- |
splicing factor-related family protein [Populus trichocarpa] |
| 2 |
Hb_002675_190 |
0.0719263336 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_001439_180 |
0.0812710852 |
- |
- |
hypothetical protein POPTR_0004s13090g [Populus trichocarpa] |
| 4 |
Hb_160818_010 |
0.0820878299 |
- |
- |
- |
| 5 |
Hb_000953_140 |
0.0875434856 |
transcription factor |
TF Family: Trihelix |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 6 |
Hb_000331_480 |
0.0950000307 |
transcription factor |
TF Family: GNAT |
PREDICTED: elongator complex protein 3 [Jatropha curcas] |
| 7 |
Hb_001034_020 |
0.0957221237 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 8 |
Hb_117950_020 |
0.0986959703 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Jatropha curcas] |
| 9 |
Hb_029281_010 |
0.0996257405 |
- |
- |
hypothetical protein JCGZ_04025 [Jatropha curcas] |
| 10 |
Hb_000491_080 |
0.1013346345 |
- |
- |
PREDICTED: F-box protein SKIP31 [Jatropha curcas] |
| 11 |
Hb_000413_130 |
0.1035754599 |
- |
- |
PREDICTED: uncharacterized protein LOC105649409 [Jatropha curcas] |
| 12 |
Hb_003734_130 |
0.1070959007 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 [Jatropha curcas] |
| 13 |
Hb_005314_010 |
0.1109170806 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 14 |
Hb_002169_010 |
0.1116329088 |
- |
- |
PREDICTED: la-related protein 6B [Jatropha curcas] |
| 15 |
Hb_000076_290 |
0.1125022681 |
- |
- |
translation factor sui1, putative [Ricinus communis] |
| 16 |
Hb_001776_080 |
0.1127796479 |
- |
- |
PREDICTED: CAX-interacting protein 4 [Jatropha curcas] |
| 17 |
Hb_011671_330 |
0.1129321756 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 4 [Jatropha curcas] |
| 18 |
Hb_002377_050 |
0.1136024451 |
- |
- |
Agmatine deiminase, putative [Ricinus communis] |
| 19 |
Hb_153547_010 |
0.114286896 |
- |
- |
PREDICTED: protein FLX-like 4 [Jatropha curcas] |
| 20 |
Hb_002149_040 |
0.1146279062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |