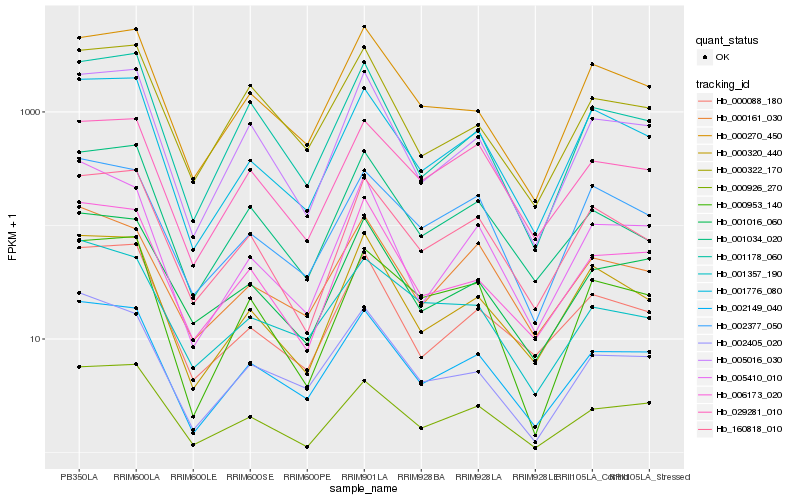
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002149_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005016_030 |
0.0745276601 |
- |
- |
hypothetical protein JCGZ_16549 [Jatropha curcas] |
| 3 |
Hb_002405_020 |
0.0769599297 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001776_080 |
0.0772956682 |
- |
- |
PREDICTED: CAX-interacting protein 4 [Jatropha curcas] |
| 5 |
Hb_001016_060 |
0.0808540318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000926_270 |
0.0860746877 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 7 |
Hb_006173_020 |
0.0897069766 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_000320_440 |
0.0925982488 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 2/3/4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001178_060 |
0.0942827852 |
- |
- |
PREDICTED: protein BUD31 homolog 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000088_180 |
0.0944026325 |
- |
- |
hypothetical protein JCGZ_11198 [Jatropha curcas] |
| 11 |
Hb_005410_010 |
0.0947938659 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07655 [Jatropha curcas] |
| 12 |
Hb_000161_030 |
0.0976443675 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP8 [Jatropha curcas] |
| 13 |
Hb_000270_450 |
0.0991888908 |
- |
- |
PREDICTED: uncharacterized protein LOC105167687 [Sesamum indicum] |
| 14 |
Hb_160818_010 |
0.1006053328 |
- |
- |
- |
| 15 |
Hb_029281_010 |
0.1009331887 |
- |
- |
hypothetical protein JCGZ_04025 [Jatropha curcas] |
| 16 |
Hb_000953_140 |
0.1027771419 |
transcription factor |
TF Family: Trihelix |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 17 |
Hb_000322_170 |
0.102839829 |
- |
- |
PREDICTED: nucleosome assembly protein 1;3 [Jatropha curcas] |
| 18 |
Hb_001034_020 |
0.1037581988 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 19 |
Hb_002377_050 |
0.1048131795 |
- |
- |
Agmatine deiminase, putative [Ricinus communis] |
| 20 |
Hb_001357_190 |
0.1058725924 |
- |
- |
ribonuclease III, putative [Ricinus communis] |