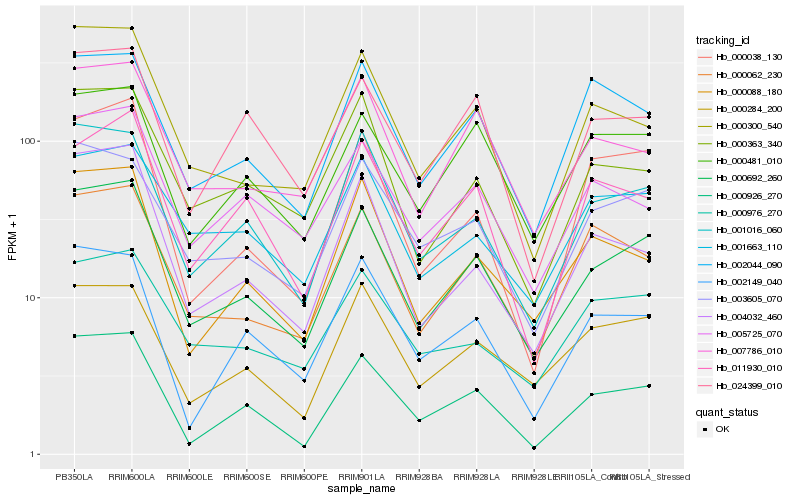
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000692_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635282 [Jatropha curcas] |
| 2 |
Hb_001016_060 |
0.08535593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000926_270 |
0.0964511027 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 4 |
Hb_003605_070 |
0.0984783783 |
- |
- |
PREDICTED: angio-associated migratory cell protein [Jatropha curcas] |
| 5 |
Hb_000284_200 |
0.1001092731 |
- |
- |
- |
| 6 |
Hb_000300_540 |
0.1027168538 |
- |
- |
PREDICTED: syntaxin-132 [Jatropha curcas] |
| 7 |
Hb_007786_010 |
0.1038535292 |
- |
- |
PREDICTED: uncharacterized protein LOC100801920 [Glycine max] |
| 8 |
Hb_000062_230 |
0.1056701742 |
- |
- |
PREDICTED: probable thimet oligopeptidase isoform X1 [Jatropha curcas] |
| 9 |
Hb_000481_010 |
0.1067430077 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP6 [Jatropha curcas] |
| 10 |
Hb_000363_340 |
0.1071891115 |
- |
- |
PREDICTED: syntaxin-52-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000088_180 |
0.1076805089 |
- |
- |
hypothetical protein JCGZ_11198 [Jatropha curcas] |
| 12 |
Hb_000976_270 |
0.1099327823 |
- |
- |
PREDICTED: uncharacterized protein LOC105646226 [Jatropha curcas] |
| 13 |
Hb_005725_070 |
0.1115851152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001663_110 |
0.1119740416 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_024399_010 |
0.1123492177 |
- |
- |
PREDICTED: uncharacterized protein LOC105649031 [Jatropha curcas] |
| 16 |
Hb_004032_460 |
0.114549643 |
- |
- |
- |
| 17 |
Hb_002149_040 |
0.1155868481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000038_130 |
0.117533596 |
- |
- |
- |
| 19 |
Hb_011930_010 |
0.1182420125 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_002044_090 |
0.1185030748 |
- |
- |
PREDICTED: uncharacterized protein LOC105643541 isoform X1 [Jatropha curcas] |