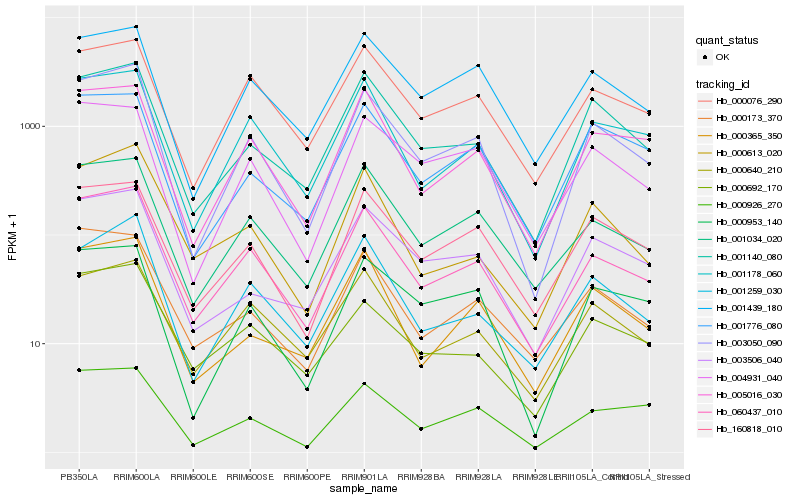
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003050_090 |
0.0 |
- |
- |
lupus la ribonucleoprotein, putative [Ricinus communis] |
| 2 |
Hb_060437_010 |
0.0728587787 |
- |
- |
E3 ubiquitin protein ligase RIE1 [Morus notabilis] |
| 3 |
Hb_001140_080 |
0.100237416 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 4 |
Hb_001034_020 |
0.1052851485 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 5 |
Hb_001178_060 |
0.1064451984 |
- |
- |
PREDICTED: protein BUD31 homolog 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000953_140 |
0.1068238344 |
transcription factor |
TF Family: Trihelix |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 7 |
Hb_001259_030 |
0.108894024 |
- |
- |
PREDICTED: uncharacterized protein LOC105641270 [Jatropha curcas] |
| 8 |
Hb_160818_010 |
0.1133944166 |
- |
- |
- |
| 9 |
Hb_000926_270 |
0.1135959026 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 10 |
Hb_005016_030 |
0.1141900367 |
- |
- |
hypothetical protein JCGZ_16549 [Jatropha curcas] |
| 11 |
Hb_003506_040 |
0.1142133398 |
- |
- |
hypothetical protein PHAVU_009G052500g [Phaseolus vulgaris] |
| 12 |
Hb_004931_040 |
0.1147283231 |
- |
- |
splicing factor-related family protein [Populus trichocarpa] |
| 13 |
Hb_000365_350 |
0.1154227417 |
- |
- |
PREDICTED: uncharacterized protein LOC105649068 [Jatropha curcas] |
| 14 |
Hb_001439_180 |
0.119706948 |
- |
- |
hypothetical protein POPTR_0004s13090g [Populus trichocarpa] |
| 15 |
Hb_001776_080 |
0.122138573 |
- |
- |
PREDICTED: CAX-interacting protein 4 [Jatropha curcas] |
| 16 |
Hb_000076_290 |
0.1239647588 |
- |
- |
translation factor sui1, putative [Ricinus communis] |
| 17 |
Hb_000692_170 |
0.1251772421 |
- |
- |
PREDICTED: uncharacterized protein LOC105638529 [Jatropha curcas] |
| 18 |
Hb_000173_370 |
0.1270180748 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 7-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000640_210 |
0.1271198156 |
- |
- |
hypothetical protein JCGZ_14045 [Jatropha curcas] |
| 20 |
Hb_000613_020 |
0.1273292081 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |