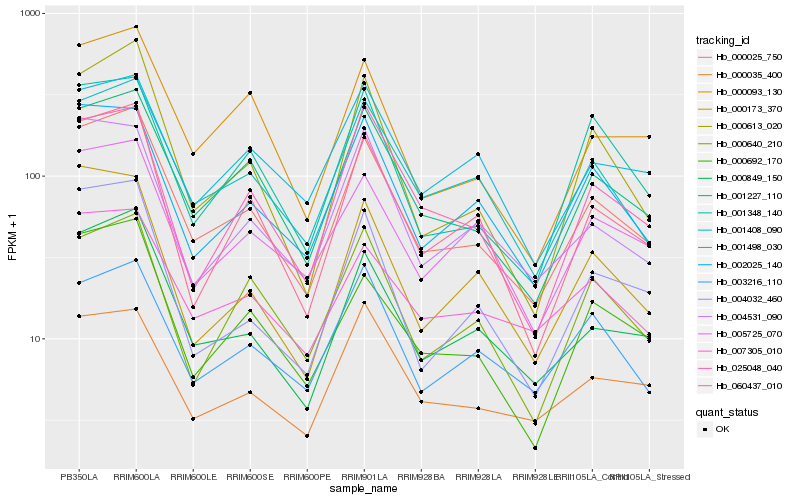
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_750 |
0.0 |
- |
- |
PREDICTED: nuclear speckle splicing regulatory protein 1-like [Jatropha curcas] |
| 2 |
Hb_001227_110 |
0.0589129509 |
- |
- |
U2 snrnp auxiliary factor, small subunit, putative [Ricinus communis] |
| 3 |
Hb_000849_150 |
0.0690386971 |
- |
- |
hypothetical protein JCGZ_15099 [Jatropha curcas] |
| 4 |
Hb_001408_090 |
0.0743663832 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 5 |
Hb_000692_170 |
0.0869493094 |
- |
- |
PREDICTED: uncharacterized protein LOC105638529 [Jatropha curcas] |
| 6 |
Hb_000093_130 |
0.0880181456 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 7 |
Hb_002025_140 |
0.0903534752 |
- |
- |
PREDICTED: UBX domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_060437_010 |
0.0906035199 |
- |
- |
E3 ubiquitin protein ligase RIE1 [Morus notabilis] |
| 9 |
Hb_007305_010 |
0.0948859394 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 10 |
Hb_000613_020 |
0.0964523685 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 11 |
Hb_005725_070 |
0.0981688206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004032_460 |
0.1035495575 |
- |
- |
- |
| 13 |
Hb_001348_140 |
0.1038993583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004531_090 |
0.1046830783 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 15 |
Hb_000640_210 |
0.1061659647 |
- |
- |
hypothetical protein JCGZ_14045 [Jatropha curcas] |
| 16 |
Hb_003216_110 |
0.1063389566 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 17 |
Hb_001498_030 |
0.1069670387 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit E [Populus euphratica] |
| 18 |
Hb_000035_400 |
0.1079989139 |
- |
- |
PREDICTED: lariat debranching enzyme-like [Citrus sinensis] |
| 19 |
Hb_025048_040 |
0.1086121834 |
- |
- |
PREDICTED: NAP1-related protein 2 [Jatropha curcas] |
| 20 |
Hb_000173_370 |
0.1096485205 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 7-like isoform X1 [Jatropha curcas] |