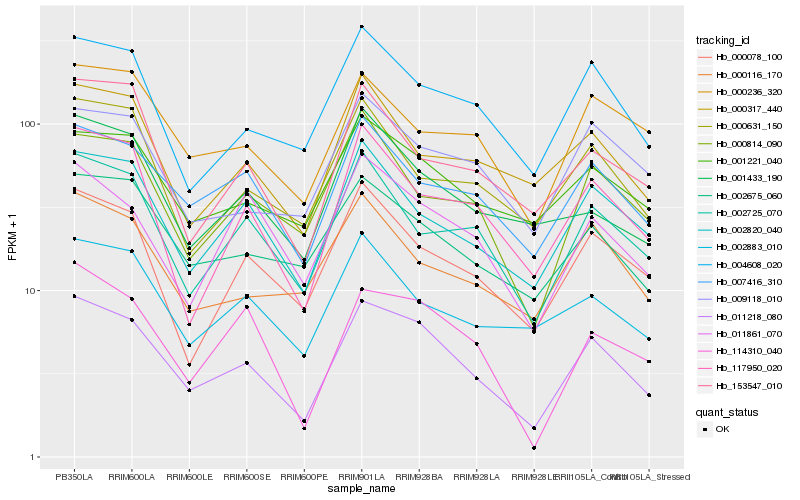
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011218_080 |
0.0 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 2 |
Hb_002820_040 |
0.1145981955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007416_310 |
0.1148545146 |
- |
- |
PREDICTED: zinc finger protein 622 [Jatropha curcas] |
| 4 |
Hb_004608_020 |
0.1174275176 |
- |
- |
PREDICTED: leucine aminopeptidase 1-like [Jatropha curcas] |
| 5 |
Hb_114310_040 |
0.1196330847 |
- |
- |
PREDICTED: purple acid phosphatase 2-like [Jatropha curcas] |
| 6 |
Hb_002675_060 |
0.1202525858 |
- |
- |
PREDICTED: uncharacterized protein LOC105634954 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002725_070 |
0.1214271056 |
- |
- |
hypothetical protein PRUPE_ppa005949mg [Prunus persica] |
| 8 |
Hb_117950_020 |
0.1218124854 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Jatropha curcas] |
| 9 |
Hb_000078_100 |
0.1223174091 |
- |
- |
PREDICTED: topless-related protein 4-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_001221_040 |
0.1275359905 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 11 |
Hb_011861_070 |
0.1275767568 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 12 |
Hb_000814_090 |
0.1310112035 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000236_320 |
0.1337961904 |
- |
- |
PREDICTED: ER membrane protein complex subunit 8/9 homolog [Jatropha curcas] |
| 14 |
Hb_000631_150 |
0.1358956506 |
- |
- |
PREDICTED: RNA polymerase-associated protein LEO1 [Jatropha curcas] |
| 15 |
Hb_000116_170 |
0.1366605727 |
- |
- |
hypothetical protein JCGZ_22392 [Jatropha curcas] |
| 16 |
Hb_153547_010 |
0.1371038857 |
- |
- |
PREDICTED: protein FLX-like 4 [Jatropha curcas] |
| 17 |
Hb_000317_440 |
0.1372083919 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 13 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001433_190 |
0.1375691963 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit A [Jatropha curcas] |
| 19 |
Hb_009118_010 |
0.1383971294 |
- |
- |
methionine aminopeptidase, putative [Ricinus communis] |
| 20 |
Hb_002883_010 |
0.1393899286 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |