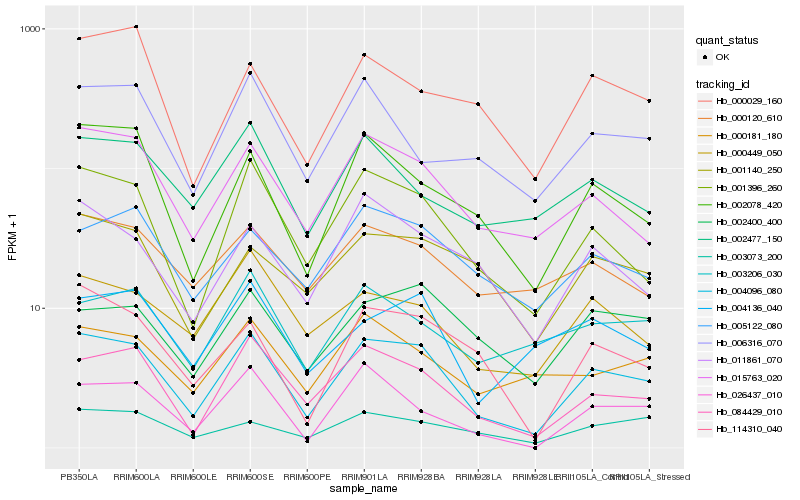
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004096_080 |
0.0 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 2 |
Hb_001396_260 |
0.1117736662 |
- |
- |
PREDICTED: F-box only protein 6 [Jatropha curcas] |
| 3 |
Hb_084429_010 |
0.1170239866 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Populus euphratica] |
| 4 |
Hb_015763_020 |
0.1325143346 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 5 |
Hb_002078_420 |
0.1388420125 |
- |
- |
PREDICTED: F-box only protein 6 [Jatropha curcas] |
| 6 |
Hb_114310_040 |
0.1425842281 |
- |
- |
PREDICTED: purple acid phosphatase 2-like [Jatropha curcas] |
| 7 |
Hb_011861_070 |
0.1524852128 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_026437_010 |
0.1564546769 |
- |
- |
- |
| 9 |
Hb_002400_400 |
0.156956939 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 10 |
Hb_000449_050 |
0.15753023 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 11 |
Hb_005122_080 |
0.1577026345 |
- |
- |
prokaryotic DNA topoisomerase, putative [Ricinus communis] |
| 12 |
Hb_003073_200 |
0.1603724491 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 13 |
Hb_001140_250 |
0.1613247608 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_004136_040 |
0.1621790058 |
- |
- |
putative protein kinase [Arabidopsis thaliana] |
| 15 |
Hb_000120_610 |
0.1637556051 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 16 |
Hb_000181_180 |
0.1638411419 |
- |
- |
PREDICTED: uncharacterized protein LOC105637608 [Jatropha curcas] |
| 17 |
Hb_000029_160 |
0.1647840935 |
- |
- |
Nitrilase, putative [Ricinus communis] |
| 18 |
Hb_003206_030 |
0.1673094311 |
- |
- |
PREDICTED: uncharacterized protein LOC105632652 [Jatropha curcas] |
| 19 |
Hb_006316_070 |
0.1674525969 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 20 |
Hb_002477_150 |
0.1680346987 |
transcription factor |
TF Family: Alfin-like |
DNA binding protein, putative [Ricinus communis] |