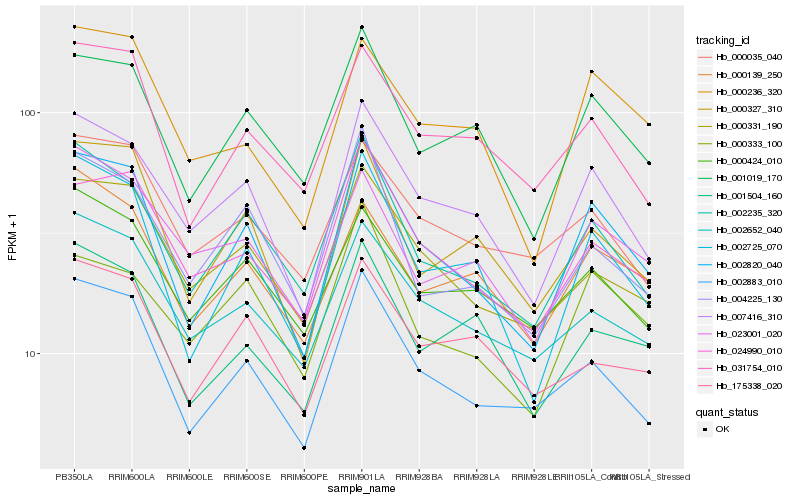
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007416_310 |
0.0 |
- |
- |
PREDICTED: zinc finger protein 622 [Jatropha curcas] |
| 2 |
Hb_002820_040 |
0.0728127816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002235_320 |
0.0737774796 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001019_170 |
0.0782706834 |
- |
- |
PREDICTED: uncharacterized protein LOC105639322 [Jatropha curcas] |
| 5 |
Hb_002652_040 |
0.0840330527 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 6 |
Hb_002725_070 |
0.0853918071 |
- |
- |
hypothetical protein PRUPE_ppa005949mg [Prunus persica] |
| 7 |
Hb_000331_190 |
0.085607599 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 8 |
Hb_024990_010 |
0.0857071269 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 9 |
Hb_000424_010 |
0.0859781692 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002883_010 |
0.0861161767 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |
| 11 |
Hb_023001_020 |
0.0869870526 |
- |
- |
PREDICTED: mitochondrial succinate-fumarate transporter 1 [Jatropha curcas] |
| 12 |
Hb_000327_310 |
0.0893279483 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 13 |
Hb_000035_040 |
0.091303501 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 14 |
Hb_000139_250 |
0.0942724238 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 15 |
Hb_000333_100 |
0.0953583478 |
- |
- |
PREDICTED: tubulin--tyrosine ligase-like protein 12 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004225_130 |
0.0958125621 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105628060 isoform X1 [Jatropha curcas] |
| 17 |
Hb_175338_020 |
0.0963241955 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 18 |
Hb_000236_320 |
0.0963936691 |
- |
- |
PREDICTED: ER membrane protein complex subunit 8/9 homolog [Jatropha curcas] |
| 19 |
Hb_001504_160 |
0.0975072024 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 20 |
Hb_031754_010 |
0.0977622382 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |