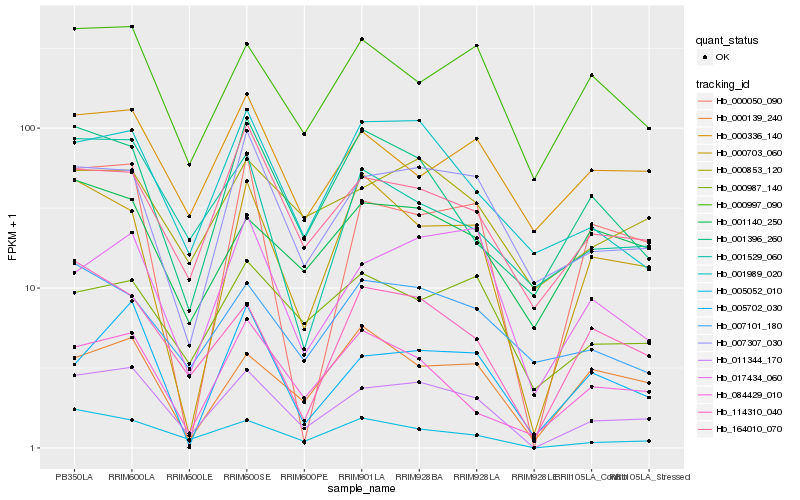
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011344_170 |
0.0 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 4-like, partial [Camelina sativa] |
| 2 |
Hb_005052_010 |
0.1426214063 |
- |
- |
PREDICTED: uncharacterized protein LOC103339315 [Prunus mume] |
| 3 |
Hb_007307_030 |
0.1460849394 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 4 |
Hb_164010_070 |
0.1526277819 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 5 |
Hb_000853_120 |
0.1620857164 |
- |
- |
PREDICTED: ethylene response sensor 1 [Jatropha curcas] |
| 6 |
Hb_017434_060 |
0.1632948038 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 7 |
Hb_001989_020 |
0.1647058797 |
- |
- |
hypothetical protein JCGZ_15521 [Jatropha curcas] |
| 8 |
Hb_000703_060 |
0.1648239742 |
- |
- |
phosphoenolpyruvate carboxylase [Manihot esculenta] |
| 9 |
Hb_114310_040 |
0.1658151633 |
- |
- |
PREDICTED: purple acid phosphatase 2-like [Jatropha curcas] |
| 10 |
Hb_000997_090 |
0.1679233739 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 11 |
Hb_084429_010 |
0.1696293733 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Populus euphratica] |
| 12 |
Hb_000987_140 |
0.174940253 |
- |
- |
PREDICTED: uncharacterized protein LOC105648083 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001140_250 |
0.1751933628 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000050_090 |
0.176792498 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 15 |
Hb_001396_260 |
0.1770505796 |
- |
- |
PREDICTED: F-box only protein 6 [Jatropha curcas] |
| 16 |
Hb_005702_030 |
0.1773465328 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 17 |
Hb_000336_140 |
0.1779753346 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007101_180 |
0.1786224863 |
- |
- |
PREDICTED: DNA polymerase beta [Jatropha curcas] |
| 19 |
Hb_000139_240 |
0.1791635663 |
- |
- |
PREDICTED: putative F-box protein At3g16210 [Jatropha curcas] |
| 20 |
Hb_001529_060 |
0.1796626061 |
- |
- |
PREDICTED: U4/U6.U5 small nuclear ribonucleoprotein 27 kDa protein [Jatropha curcas] |