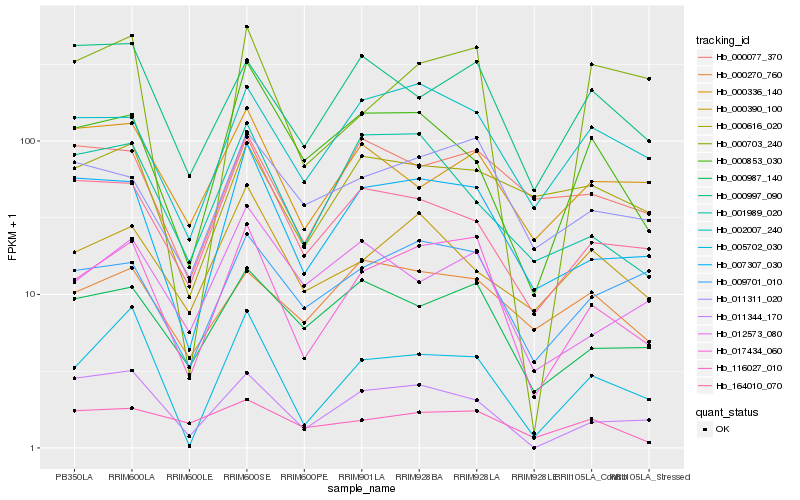
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017434_060 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 2 |
Hb_007307_030 |
0.1262317 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 3 |
Hb_011311_020 |
0.1536872224 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 4 |
Hb_005702_030 |
0.154101441 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 5 |
Hb_002007_240 |
0.1579754165 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 6 |
Hb_000987_140 |
0.1590193748 |
- |
- |
PREDICTED: uncharacterized protein LOC105648083 isoform X1 [Jatropha curcas] |
| 7 |
Hb_011344_170 |
0.1632948038 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 4-like, partial [Camelina sativa] |
| 8 |
Hb_009701_010 |
0.166107974 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 9 |
Hb_164010_070 |
0.1808163438 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 10 |
Hb_000997_090 |
0.1870569863 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 11 |
Hb_012573_080 |
0.1892542424 |
- |
- |
hypothetical protein JCGZ_13311 [Jatropha curcas] |
| 12 |
Hb_000077_370 |
0.1892662112 |
- |
- |
PREDICTED: protein STABILIZED1 [Jatropha curcas] |
| 13 |
Hb_001989_020 |
0.1908422673 |
- |
- |
hypothetical protein JCGZ_15521 [Jatropha curcas] |
| 14 |
Hb_000390_100 |
0.1918217292 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 15 |
Hb_000853_030 |
0.1924083467 |
- |
- |
sucrose synthase 2 [Hevea brasiliensis] |
| 16 |
Hb_000703_240 |
0.1932624572 |
- |
- |
- |
| 17 |
Hb_000616_020 |
0.1939905556 |
- |
- |
PREDICTED: cysteine desulfurase, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_000270_760 |
0.1953463441 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000336_140 |
0.1979442183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_116027_010 |
0.1980468276 |
- |
- |
PREDICTED: separase [Jatropha curcas] |