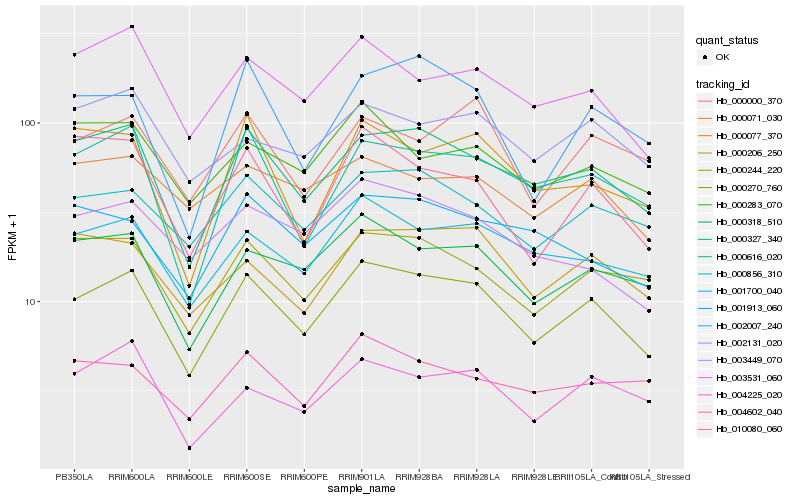
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_760 |
0.0 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000327_340 |
0.0706027201 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 3 |
Hb_000318_510 |
0.0857757356 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |
| 4 |
Hb_001700_040 |
0.0865136605 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex non-core subunit NAF1 [Jatropha curcas] |
| 5 |
Hb_000244_220 |
0.0909773535 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000856_310 |
0.0910200849 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 7 |
Hb_003531_060 |
0.0961085713 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_000206_250 |
0.097653886 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 9 |
Hb_000616_020 |
0.0983691008 |
- |
- |
PREDICTED: cysteine desulfurase, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_004225_020 |
0.1064153136 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000000_370 |
0.1070732699 |
transcription factor |
TF Family: BES1 |
PREDICTED: protein BRASSINAZOLE-RESISTANT 1 [Jatropha curcas] |
| 12 |
Hb_002007_240 |
0.1086899278 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 13 |
Hb_000283_070 |
0.1087705335 |
- |
- |
PREDICTED: T-complex protein 1 subunit delta [Jatropha curcas] |
| 14 |
Hb_002131_020 |
0.1089771985 |
transcription factor |
TF Family: IWS1 |
PREDICTED: protein IWS1 homolog [Jatropha curcas] |
| 15 |
Hb_004602_040 |
0.1091814298 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_003449_070 |
0.1095471886 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |
| 17 |
Hb_010080_060 |
0.1099902081 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_000071_030 |
0.1107200185 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT31 [Jatropha curcas] |
| 19 |
Hb_001913_060 |
0.1110523849 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 20 |
Hb_000077_370 |
0.1116169616 |
- |
- |
PREDICTED: protein STABILIZED1 [Jatropha curcas] |