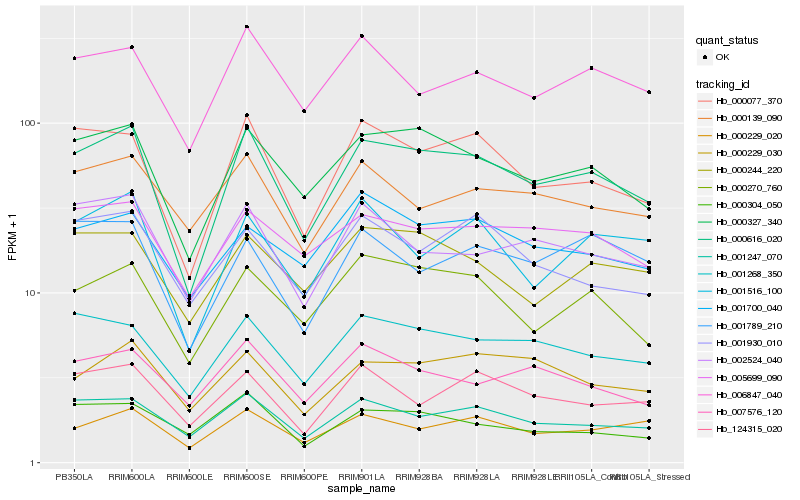
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000616_020 |
0.0 |
- |
- |
PREDICTED: cysteine desulfurase, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_000327_340 |
0.0686743094 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 3 |
Hb_000077_370 |
0.0740307234 |
- |
- |
PREDICTED: protein STABILIZED1 [Jatropha curcas] |
| 4 |
Hb_001247_070 |
0.0937085946 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g56690, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001268_350 |
0.0942018044 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105115846 [Populus euphratica] |
| 6 |
Hb_000270_760 |
0.0983691008 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006847_040 |
0.1067963596 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 1 [Jatropha curcas] |
| 8 |
Hb_005699_090 |
0.1079561442 |
- |
- |
PREDICTED: uncharacterized protein LOC105639563 [Jatropha curcas] |
| 9 |
Hb_001516_100 |
0.1096831026 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_002524_040 |
0.1099190626 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 11 |
Hb_000244_220 |
0.1102276065 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000229_030 |
0.1111846572 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g73710 [Jatropha curcas] |
| 13 |
Hb_000229_020 |
0.1113470731 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03540 [Jatropha curcas] |
| 14 |
Hb_124315_020 |
0.1141230506 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_001789_210 |
0.1151513639 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000139_090 |
0.1157028509 |
- |
- |
PREDICTED: protein TIME FOR COFFEE isoform X3 [Jatropha curcas] |
| 17 |
Hb_007576_120 |
0.1162898382 |
- |
- |
NBS-LRR disease resistance protein NBS49 [Dimocarpus longan] |
| 18 |
Hb_001700_040 |
0.1194571256 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex non-core subunit NAF1 [Jatropha curcas] |
| 19 |
Hb_000304_050 |
0.1200811682 |
transcription factor |
TF Family: GRAS |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 20 |
Hb_001930_010 |
0.1205101616 |
- |
- |
PREDICTED: uncharacterized protein LOC105633074 [Jatropha curcas] |