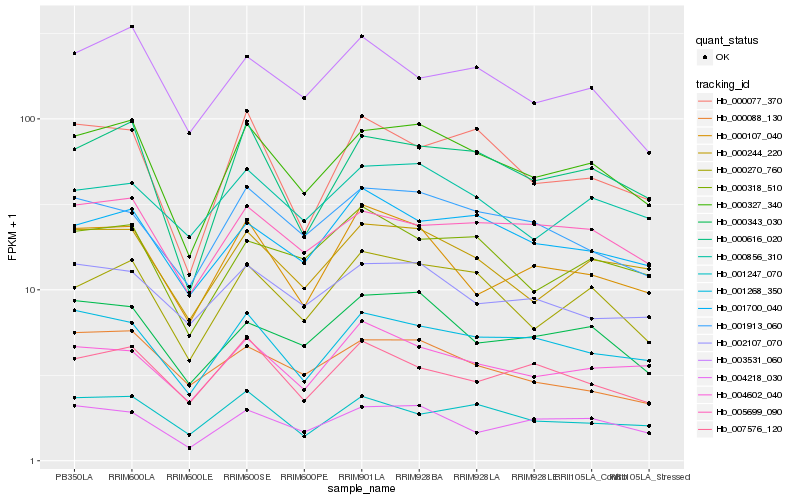
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000327_340 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 2 |
Hb_000616_020 |
0.0686743094 |
- |
- |
PREDICTED: cysteine desulfurase, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_000270_760 |
0.0706027201 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000244_220 |
0.0772973595 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_001268_350 |
0.0828323069 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105115846 [Populus euphratica] |
| 6 |
Hb_005699_090 |
0.0848287866 |
- |
- |
PREDICTED: uncharacterized protein LOC105639563 [Jatropha curcas] |
| 7 |
Hb_000343_030 |
0.0871344897 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 8 |
Hb_004218_030 |
0.0886992772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001913_060 |
0.0899141558 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 10 |
Hb_001700_040 |
0.0982137811 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex non-core subunit NAF1 [Jatropha curcas] |
| 11 |
Hb_000856_310 |
0.0989277491 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 12 |
Hb_000318_510 |
0.0998890379 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |
| 13 |
Hb_000077_370 |
0.100294504 |
- |
- |
PREDICTED: protein STABILIZED1 [Jatropha curcas] |
| 14 |
Hb_002107_070 |
0.100775302 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 15 |
Hb_001247_070 |
0.1008901349 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g56690, mitochondrial [Jatropha curcas] |
| 16 |
Hb_003531_060 |
0.1014527174 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_000107_040 |
0.1034518944 |
- |
- |
nuclear pore complex protein nup153, putative [Ricinus communis] |
| 18 |
Hb_004602_040 |
0.1053628503 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_007576_120 |
0.1066666643 |
- |
- |
NBS-LRR disease resistance protein NBS49 [Dimocarpus longan] |
| 20 |
Hb_000088_130 |
0.1069186201 |
- |
- |
hypothetical protein JCGZ_02447 [Jatropha curcas] |