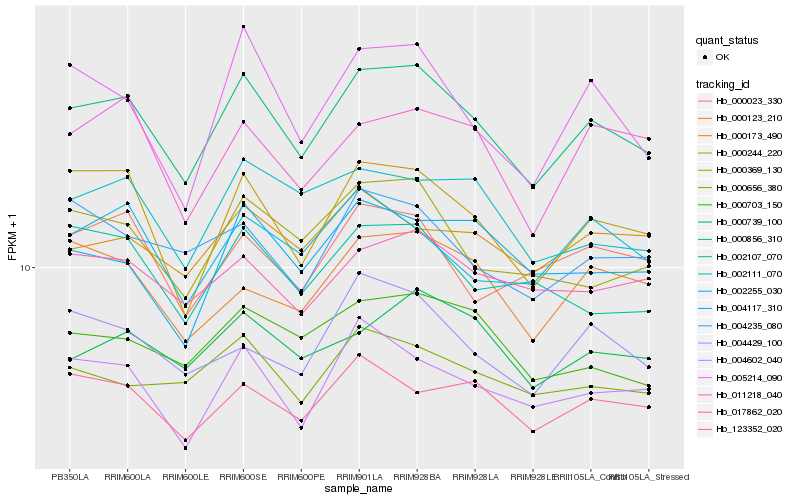
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000856_310 |
0.0 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 2 |
Hb_000244_220 |
0.0638155194 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_004602_040 |
0.077895006 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_005214_090 |
0.0787800794 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 5 |
Hb_000369_130 |
0.0793029183 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 6 |
Hb_017862_020 |
0.0797782985 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase lvsG [Jatropha curcas] |
| 7 |
Hb_000739_100 |
0.0801269329 |
- |
- |
ku P80 DNA helicase, putative [Ricinus communis] |
| 8 |
Hb_123352_020 |
0.0804584065 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g65560 [Jatropha curcas] |
| 9 |
Hb_000703_150 |
0.0806088067 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A [Jatropha curcas] |
| 10 |
Hb_000656_380 |
0.0812851964 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 11 |
Hb_011218_040 |
0.0831390483 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_002111_070 |
0.0838537931 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 13 |
Hb_000123_210 |
0.0840328528 |
- |
- |
PREDICTED: uncharacterized protein LOC105641576 isoform X2 [Jatropha curcas] |
| 14 |
Hb_004429_100 |
0.0857624224 |
- |
- |
PREDICTED: asparagine synthetase domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_002255_030 |
0.086113619 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 16 |
Hb_000023_330 |
0.0862189226 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 17 |
Hb_004117_310 |
0.0863120384 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 18 |
Hb_002107_070 |
0.0867646977 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 19 |
Hb_004235_080 |
0.0867783007 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 20 |
Hb_000173_490 |
0.0874644566 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 isoform X2 [Jatropha curcas] |