| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
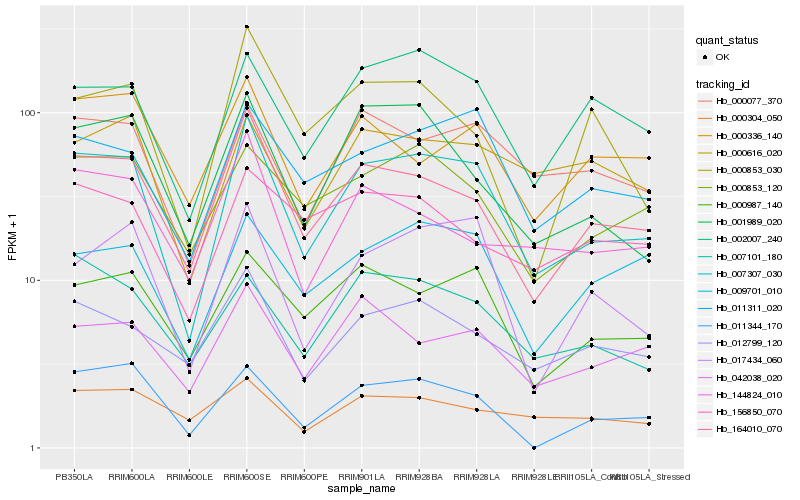
Hb_007307_030 |
0.0 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 2 |
Hb_164010_070 |
0.1088849118 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 3 |
Hb_017434_060 |
0.1262317 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 4 |
Hb_011311_020 |
0.1267937977 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000077_370 |
0.1323444643 |
- |
- |
PREDICTED: protein STABILIZED1 [Jatropha curcas] |
| 6 |
Hb_000987_140 |
0.1344779359 |
- |
- |
PREDICTED: uncharacterized protein LOC105648083 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002007_240 |
0.1351874912 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 8 |
Hb_001989_020 |
0.1374043994 |
- |
- |
hypothetical protein JCGZ_15521 [Jatropha curcas] |
| 9 |
Hb_012799_120 |
0.141048572 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_009701_010 |
0.1441940589 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 11 |
Hb_007101_180 |
0.1449915285 |
- |
- |
PREDICTED: DNA polymerase beta [Jatropha curcas] |
| 12 |
Hb_042038_020 |
0.1450053982 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 13 |
Hb_011344_170 |
0.1460849394 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 4-like, partial [Camelina sativa] |
| 14 |
Hb_000853_120 |
0.1465852764 |
- |
- |
PREDICTED: ethylene response sensor 1 [Jatropha curcas] |
| 15 |
Hb_000336_140 |
0.1538990197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000616_020 |
0.1555938498 |
- |
- |
PREDICTED: cysteine desulfurase, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_144824_010 |
0.1562728617 |
- |
- |
hypothetical protein POPTR_0019s02110g [Populus trichocarpa] |
| 18 |
Hb_000304_050 |
0.1615035793 |
transcription factor |
TF Family: GRAS |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 19 |
Hb_156850_070 |
0.1632681909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000853_030 |
0.163323646 |
- |
- |
sucrose synthase 2 [Hevea brasiliensis] |