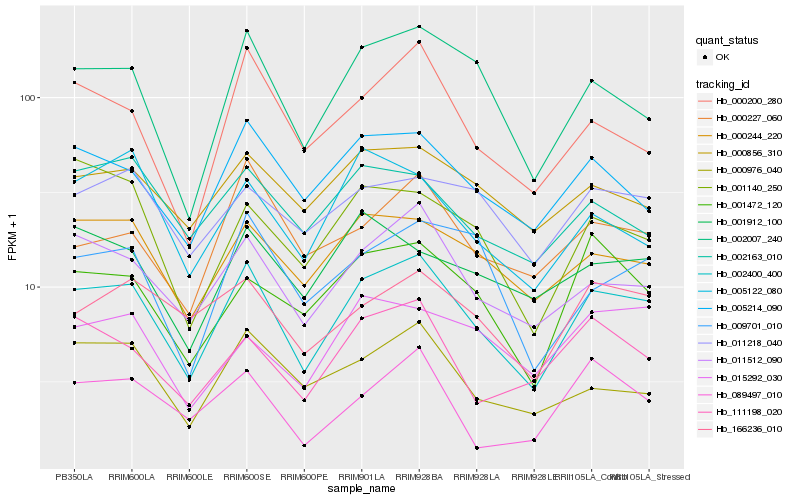
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002400_400 |
0.0 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 2 |
Hb_011512_090 |
0.0996834184 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 3 |
Hb_002007_240 |
0.1030028796 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 4 |
Hb_000244_220 |
0.1099517538 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_005214_090 |
0.1102743163 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 6 |
Hb_000200_280 |
0.1183949828 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 7 |
Hb_000976_040 |
0.1215839021 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 8 |
Hb_166236_010 |
0.1237567695 |
- |
- |
- |
| 9 |
Hb_001472_120 |
0.1237874538 |
- |
- |
- |
| 10 |
Hb_000856_310 |
0.1245483293 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 11 |
Hb_111198_020 |
0.12572949 |
- |
- |
hypothetical protein JCGZ_06907 [Jatropha curcas] |
| 12 |
Hb_000227_060 |
0.1292259354 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 13 |
Hb_011218_040 |
0.1298490263 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_009701_010 |
0.1301152562 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 15 |
Hb_002163_010 |
0.1320225696 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 16 |
Hb_015292_030 |
0.1363093296 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02490, mitochondrial [Jatropha curcas] |
| 17 |
Hb_001912_100 |
0.1367877515 |
- |
- |
PREDICTED: uncharacterized protein LOC105636609 [Jatropha curcas] |
| 18 |
Hb_001140_250 |
0.1371998511 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_005122_080 |
0.137227297 |
- |
- |
prokaryotic DNA topoisomerase, putative [Ricinus communis] |
| 20 |
Hb_089497_010 |
0.1380497156 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |