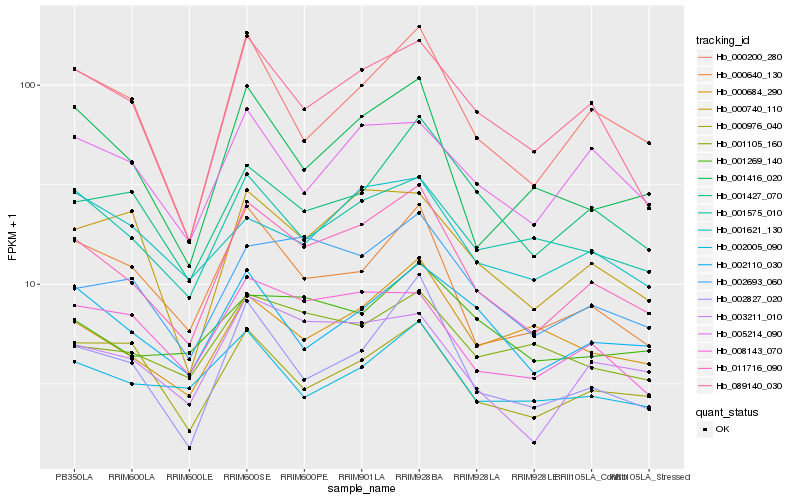
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011716_090 |
0.0 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 2 |
Hb_089140_030 |
0.098382258 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 3 |
Hb_000200_280 |
0.1005865143 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 4 |
Hb_002693_060 |
0.1033358896 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_000640_130 |
0.1058072761 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_002827_020 |
0.1072437606 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 7 |
Hb_003211_010 |
0.1080237876 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 8 |
Hb_001416_020 |
0.1127344705 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 9 |
Hb_008143_070 |
0.1134730728 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 10 |
Hb_001575_010 |
0.1136863607 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000740_110 |
0.1181609223 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01850 [Jatropha curcas] |
| 12 |
Hb_002110_030 |
0.1190649005 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 13 |
Hb_001427_070 |
0.1193845572 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 14 |
Hb_005214_090 |
0.1217052563 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 15 |
Hb_000684_290 |
0.1217173858 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 16 |
Hb_002005_090 |
0.1240359093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000976_040 |
0.1245064624 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 18 |
Hb_001269_140 |
0.1256978627 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 19 |
Hb_001621_130 |
0.1282700392 |
- |
- |
PREDICTED: pumilio homolog 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001105_160 |
0.128675049 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |