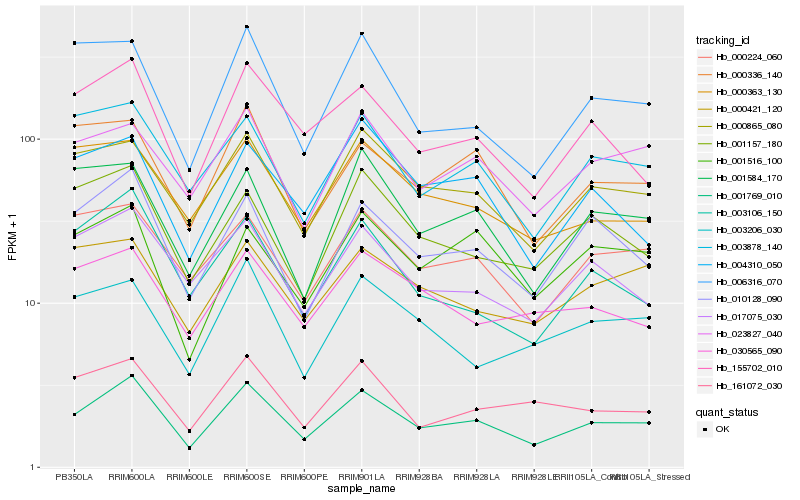
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001769_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_010128_090 |
0.0961890053 |
- |
- |
PREDICTED: uncharacterized protein LOC105647377 [Jatropha curcas] |
| 3 |
Hb_000865_080 |
0.0984835762 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 4 |
Hb_155702_010 |
0.1055449077 |
- |
- |
Tetraspanin-5 [Gossypium arboreum] |
| 5 |
Hb_023827_040 |
0.1087383561 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 6 |
Hb_003878_140 |
0.1108888235 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 7 |
Hb_000224_060 |
0.1128789827 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6 [Jatropha curcas] |
| 8 |
Hb_017075_030 |
0.113632918 |
- |
- |
helicase, putative [Ricinus communis] |
| 9 |
Hb_000336_140 |
0.113879788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001516_100 |
0.1161648901 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_006316_070 |
0.1163716372 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 12 |
Hb_001584_170 |
0.1166267103 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 13 |
Hb_161072_030 |
0.1177644371 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77405 [Jatropha curcas] |
| 14 |
Hb_001157_180 |
0.117852484 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_003106_150 |
0.1201735061 |
- |
- |
PREDICTED: U-box domain-containing protein 38-like [Jatropha curcas] |
| 16 |
Hb_004310_050 |
0.122357504 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 17 |
Hb_000363_130 |
0.122497636 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |
| 18 |
Hb_000421_120 |
0.1236324383 |
- |
- |
PREDICTED: NEDD8-specific protease 1 [Jatropha curcas] |
| 19 |
Hb_003206_030 |
0.1238129901 |
- |
- |
PREDICTED: uncharacterized protein LOC105632652 [Jatropha curcas] |
| 20 |
Hb_030565_090 |
0.1242736333 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |