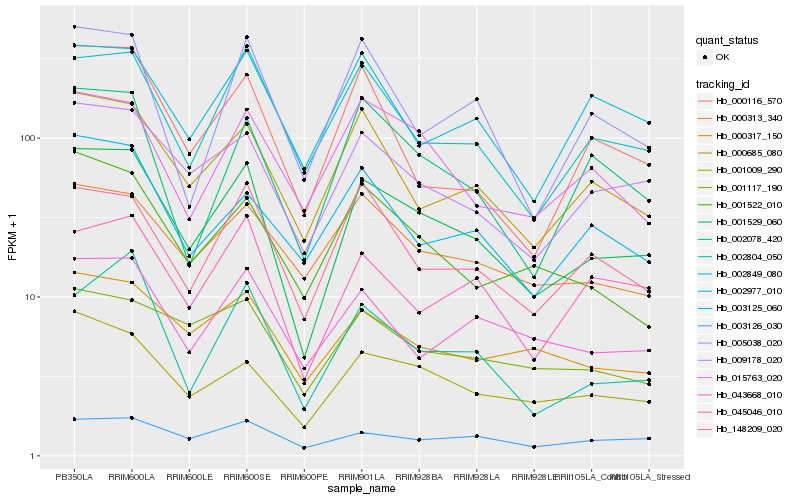
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001529_060 |
0.0 |
- |
- |
PREDICTED: U4/U6.U5 small nuclear ribonucleoprotein 27 kDa protein [Jatropha curcas] |
| 2 |
Hb_009178_020 |
0.1034209271 |
- |
- |
PREDICTED: zinc finger protein 622 [Jatropha curcas] |
| 3 |
Hb_001009_290 |
0.1128383584 |
- |
- |
PREDICTED: elongator complex protein 3-like [Populus euphratica] |
| 4 |
Hb_000685_080 |
0.1162611613 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_002849_080 |
0.1176403721 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 6 |
Hb_148209_020 |
0.1261106004 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 7 |
Hb_000317_150 |
0.127286695 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_045046_010 |
0.1273775506 |
transcription factor |
TF Family: TUB |
hypothetical protein POPTR_0011s11070g [Populus trichocarpa] |
| 9 |
Hb_002078_420 |
0.1298630159 |
- |
- |
PREDICTED: F-box only protein 6 [Jatropha curcas] |
| 10 |
Hb_002804_050 |
0.131252553 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 11 |
Hb_003126_030 |
0.1316039867 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000313_340 |
0.1329372143 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 13 |
Hb_043668_010 |
0.1332500615 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_015763_020 |
0.1337494317 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 15 |
Hb_001522_010 |
0.1350743159 |
- |
- |
PREDICTED: nucleolar protein 14 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001117_190 |
0.1367669371 |
transcription factor |
TF Family: FAR1 |
FAR1-related sequence 11 [Theobroma cacao] |
| 17 |
Hb_005038_020 |
0.1375243513 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_002977_010 |
0.1391005975 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 19 |
Hb_003125_060 |
0.1396634527 |
- |
- |
PREDICTED: snurportin-1 [Jatropha curcas] |
| 20 |
Hb_000116_570 |
0.1399970153 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |