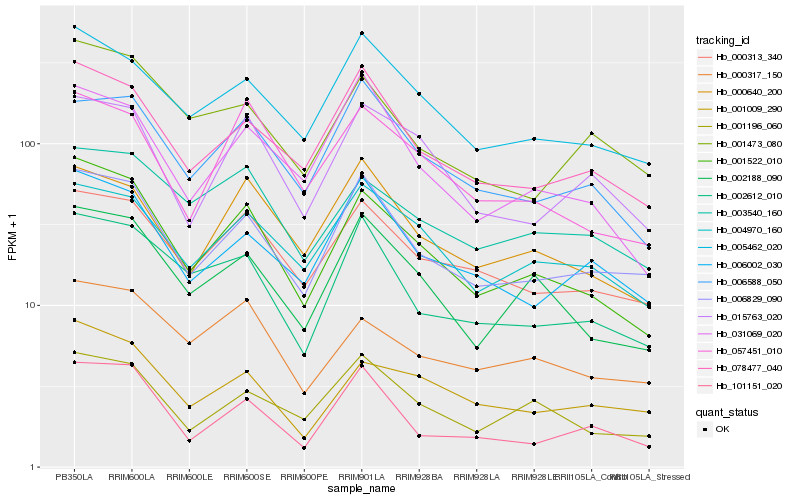
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001522_010 |
0.0 |
- |
- |
PREDICTED: nucleolar protein 14 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002188_090 |
0.0876560064 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X1 [Jatropha curcas] |
| 3 |
Hb_006829_090 |
0.0999824723 |
- |
- |
PREDICTED: protein MOS2 [Jatropha curcas] |
| 4 |
Hb_078477_040 |
0.1028718866 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 5 |
Hb_005462_020 |
0.1032485119 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 6 |
Hb_001009_290 |
0.1066423794 |
- |
- |
PREDICTED: elongator complex protein 3-like [Populus euphratica] |
| 7 |
Hb_000313_340 |
0.1069386868 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 8 |
Hb_002612_010 |
0.1081704262 |
- |
- |
hypothetical protein JCGZ_19269 [Jatropha curcas] |
| 9 |
Hb_057451_010 |
0.1082014404 |
- |
- |
PREDICTED: uncharacterized protein LOC105635588 [Jatropha curcas] |
| 10 |
Hb_000317_150 |
0.1100788943 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_001196_060 |
0.1111567827 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 12 |
Hb_004970_160 |
0.1174880673 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_003540_160 |
0.1175867185 |
- |
- |
hypothetical protein POPTR_0011s12160g [Populus trichocarpa] |
| 14 |
Hb_000640_200 |
0.1182158983 |
- |
- |
hypothetical protein RCOM_1346600 [Ricinus communis] |
| 15 |
Hb_006002_030 |
0.1183801862 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 16 |
Hb_015763_020 |
0.1195697097 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 17 |
Hb_031069_020 |
0.1197249393 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 3-like [Jatropha curcas] |
| 18 |
Hb_001473_080 |
0.1206755901 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 19 |
Hb_101151_020 |
0.1220435922 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_006588_050 |
0.1224794589 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |