| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
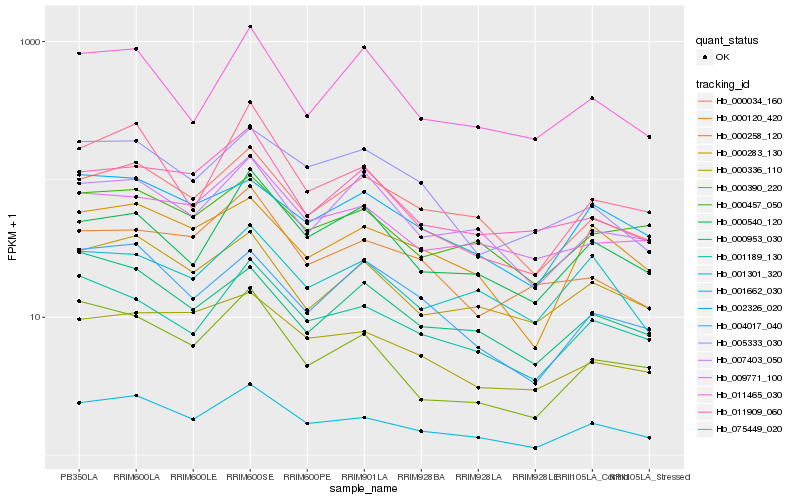
Hb_001662_030 |
0.0 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Populus euphratica] |
| 2 |
Hb_000034_160 |
0.0983653044 |
- |
- |
hypothetical protein EUGRSUZ_F02151 [Eucalyptus grandis] |
| 3 |
Hb_000258_120 |
0.1008066969 |
- |
- |
PREDICTED: uncharacterized protein LOC105630440 [Jatropha curcas] |
| 4 |
Hb_001189_130 |
0.1016835482 |
transcription factor |
TF Family: NAC |
PREDICTED: protein ATAF2-like [Jatropha curcas] |
| 5 |
Hb_075449_020 |
0.1038250094 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 6 |
Hb_000390_220 |
0.1039111067 |
- |
- |
PREDICTED: uncharacterized protein LOC100244219 [Vitis vinifera] |
| 7 |
Hb_000283_130 |
0.1050928545 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 8 |
Hb_000336_110 |
0.11130721 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_004017_040 |
0.1132587788 |
- |
- |
kinetochore protein nuf2, putative [Ricinus communis] |
| 10 |
Hb_000457_050 |
0.1197723154 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 11 |
Hb_002326_020 |
0.1199208934 |
- |
- |
hypothetical protein POPTR_0002s13160g [Populus trichocarpa] |
| 12 |
Hb_007403_050 |
0.1212174232 |
- |
- |
PREDICTED: protein YIPF5 homolog [Jatropha curcas] |
| 13 |
Hb_005333_030 |
0.1223539395 |
- |
- |
hypothetical protein JCGZ_15866 [Jatropha curcas] |
| 14 |
Hb_009771_100 |
0.1227677766 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 15 |
Hb_011909_060 |
0.1238971974 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 16 |
Hb_000540_120 |
0.1239927489 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 17 |
Hb_000953_030 |
0.1269776224 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000120_420 |
0.1284486113 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 19 |
Hb_011465_030 |
0.1295086147 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 20 |
Hb_001301_320 |
0.1303180282 |
- |
- |
PREDICTED: uncharacterized protein LOC105632955 [Jatropha curcas] |