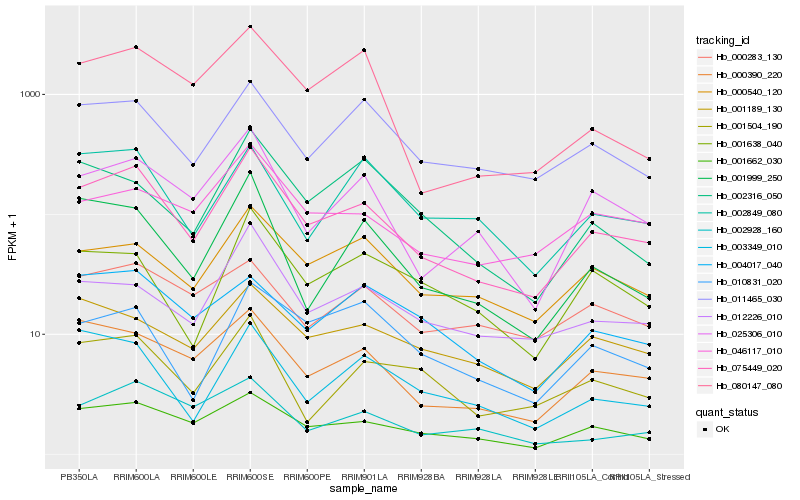
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_075449_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 2 |
Hb_001662_030 |
0.1038250094 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Populus euphratica] |
| 3 |
Hb_000390_220 |
0.1159442878 |
- |
- |
PREDICTED: uncharacterized protein LOC100244219 [Vitis vinifera] |
| 4 |
Hb_025306_010 |
0.1223450824 |
transcription factor |
TF Family: MYB-related |
syringolide-induced protein 1-3-1B [Populus trichocarpa] |
| 5 |
Hb_000540_120 |
0.1316911516 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 6 |
Hb_011465_030 |
0.132993757 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 7 |
Hb_001999_250 |
0.1349076607 |
- |
- |
PREDICTED: uncharacterized protein LOC105133073 isoform X1 [Populus euphratica] |
| 8 |
Hb_046117_010 |
0.1366552345 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 9 |
Hb_012226_010 |
0.1402573544 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 10 |
Hb_001189_130 |
0.1450121418 |
transcription factor |
TF Family: NAC |
PREDICTED: protein ATAF2-like [Jatropha curcas] |
| 11 |
Hb_001504_190 |
0.1456479483 |
- |
- |
double-stranded RNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_003349_010 |
0.1465478523 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CICLE_v10023911mg [Citrus clementina] |
| 13 |
Hb_001638_040 |
0.147067477 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 14 |
Hb_002928_160 |
0.1474368279 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 15 |
Hb_080147_080 |
0.1490750504 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 16 |
Hb_002849_080 |
0.149590665 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 17 |
Hb_010831_020 |
0.1535733609 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 18 |
Hb_004017_040 |
0.1552568275 |
- |
- |
kinetochore protein nuf2, putative [Ricinus communis] |
| 19 |
Hb_002316_050 |
0.1565075571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000283_130 |
0.156517793 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |